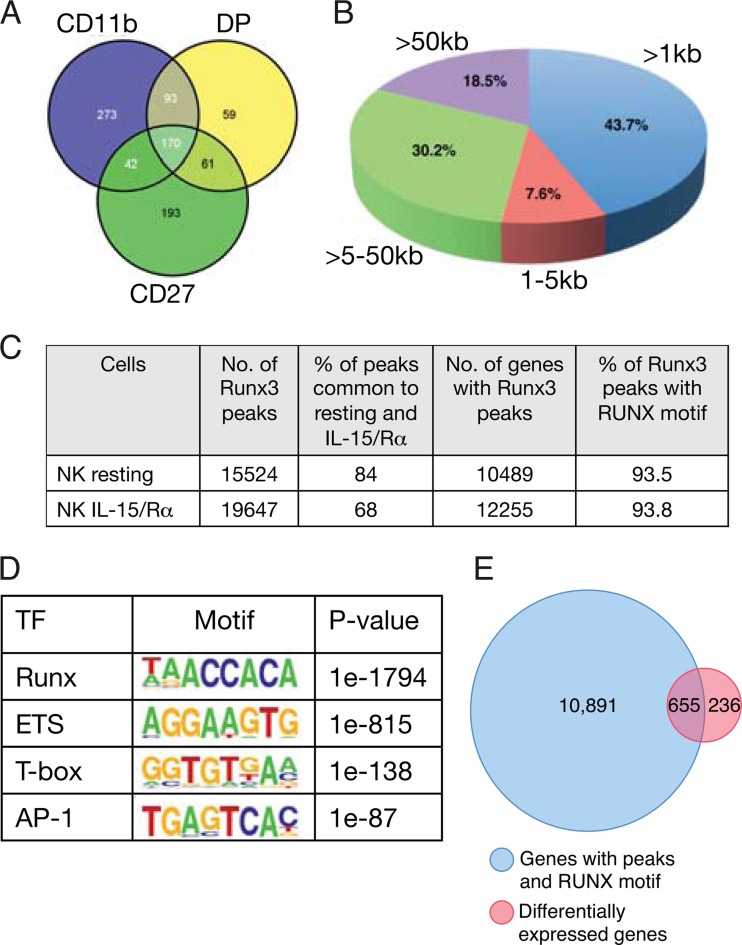
FIG 6.
Integrated analysis of Runx3 ChIP-seq and gene expression data of in vivo IL-15-activated NKC. (A) Intersection of differentially expressed genes (Runx3−/−/WT ≥ 1.5-fold; P < 0.05) in the three spleen NKC subpopulations of WT and Runx3−/− mice (CD27, CD27+ CD11b−; DP, CD27+ CD11b+; CD11b, CD11b+ CD27−). (B) Pie chart showing the genomic distribution of Runx3 peaks in in vivo IL-15/Rα-activated NKC. (C) Analysis of Runx3 peaks in NKC in vivo. Runx3 peak numbers, corresponding genes, and percentages of peaks with RUNX motif (RCCRCA) in resting NK cells (68) and in in vivo IL-15-activated NKC. (D) De novo-discovered enriched TF motifs in Runx3 peaks in in vivo IL-15/Rα-activated NKC. (E) In vivo IL-15-activated Runx3-regulated genes: intersection between the list of genes containing Runx3 peaks with a RUNX motif and the list of differentially expressed genes (Runx3−/−/WT ≥ 1.5-fold; P < 0.05) in the three spleen NKC populations.