FIG 3.
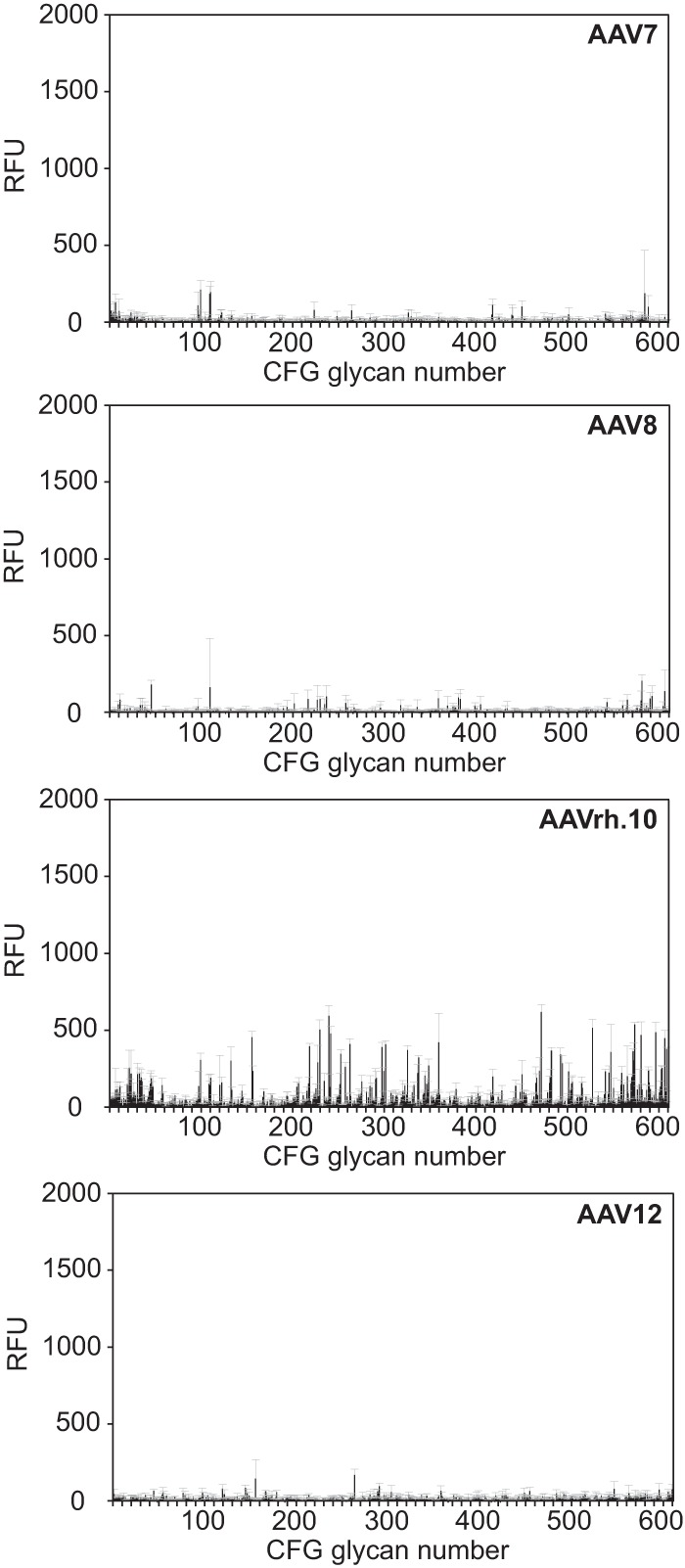
CFG glycan array screens of AAV7, -8, -rh.10, and -12. Shown are binding efficiencies of fluorescently labeled AAV7, -8, -rh.10, and -12 vectors on CFG glycan arrays displaying 611 different glycan structures, each in replicates of six. Note that binding signals for glycans 81, 265, and 518 were removed from the AAV7 and -rh.10 screens, respectively, based on a statement of the CFG that these glycans repeatedly produced nonspecific signals in unrelated glycan arrays performed with the same array batch. Error bars represent standard deviations.
