FIG 3.
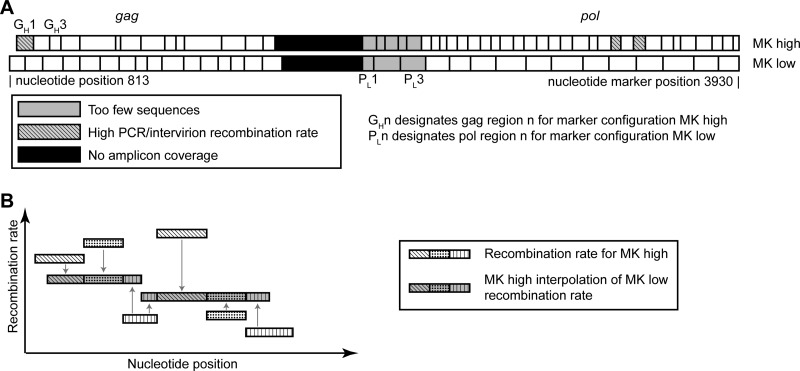
Schematic of marker configurations and how to compare between them. (A) In this study, recombination is measured between wild-type virus and a marker system with silent codon modification markers that do not affect any viral proteins or packaging (marker configuration MKhigh). To test that these codon modifications do not influence our recombination rate measurements, a second marker system virus is created where the codon modifications occur at different nucleotide positions (marker configuration MKlow). (B) To compare between marker configurations, MKhigh is used to predict what would be measured as the recombination rate if MKlow was used. This prediction can then be directly compared to the experimental results for MKlow. For each interval in MKlow, the MKhigh prediction is calculated by averaging the overlapping MKhigh interval's recombination rate and weighting this average by the proportion of overlap.