ABSTRACT
Arabidopsis encodes five double-stranded RNA binding (DRB) proteins. DRB1 and DRB2 are involved in microRNA (miRNA) biogenesis, while DRB4 functions in cytoplasmic posttranscriptional small interfering RNA (siRNA) pathways. DRB3 and DRB5 are not involved in double-stranded RNA (dsRNA) processing but assist in silencing transcripts targeted by DRB2-associated miRNAs. The goal of this study was to determine which, if any, of the DRB proteins might also participate in a nuclear siRNA pathway that leads to geminivirus genome methylation. Here, we demonstrate that DRB3 functions with Dicer-like 3 (DCL3) and Argonaute 4 (AGO4) in methylation-mediated antiviral defense. Plants employ repressive viral genome methylation as an epigenetic defense against geminiviruses, using an RNA-directed DNA methylation (RdDM) pathway similar to that used to suppress endogenous invasive DNAs such as transposons. Chromatin methylation inhibits virus replication and transcription, and methylation-deficient host plants are hypersusceptible to geminivirus infection. Using a panel of drb mutants, we found that drb3 plants uniquely exhibit a similar hypersensitivity and that viral genome methylation is substantially reduced in drb3 compared to wild-type plants. In addition, like dcl3 and ago4 mutants, drb3 plants fail to recover from infection and cannot accomplish the viral genome hypermethylation that is invariably observed in asymptomatic, recovered tissues. Small RNA analysis, bimolecular fluorescence complementation, and coimmunoprecipitation experiments show that DRB3 acts downstream of siRNA biogenesis and suggest that it associates with DCL3 and AGO4 in distinct subnuclear compartments. These studies reveal that in addition to its previously established role in the miRNA pathway, DRB3 also functions in antiviral RdDM.
IMPORTANCE Plants use RNA-directed DNA methylation (RdDM) as an epigenetic defense against geminiviruses. RNA silencing pathways in Arabidopsis include five double-stranded RNA binding proteins (DRBs) related to Drosophila R2D2 and mammalian TRBP and PACT. While DRB proteins have defined roles in miRNA and cytoplasmic siRNA pathways, a role in nuclear RdDM was elusive. Here, we used the geminivirus system to show that DRB3 is involved in methylation-mediated antiviral defense. Beginning with a panel of Arabidopsis drb mutants, we demonstrated that drb3 plants uniquely show enhanced susceptibility to geminiviruses. Further, like dcl3 and ago4 mutants, drb3 plants fail to hypermethylate the viral genome, a requirement for host recovery. We also show that DRB3 physically interacts with the RdDM pathway components DCL3 and AGO4 in the nucleus. This work highlights the utility of geminiviruses as models for de novo RdDM and places DRB3 protein in this fundamental epigenetic pathway.
INTRODUCTION
Plants use a sophisticated RNA silencing machinery to control the expression of selected genes and repress resident transposons. Silencing can occur posttranscriptionally through several pathways mediated by microRNA (miRNA) or small interfering RNA (siRNA) or transcriptionally by small RNA-directed DNA methylation (RdDM) (1–6). In addition, RNA silencing acts as an effective defense against invading nucleic acids, including those of RNA and DNA viruses (7–9). The small RNAs characteristic of individual silencing pathways are generated by distinct Dicer-like (DCL) ribonucleases that process larger double-stranded RNA (dsRNA) and hairpin precursors. In Arabidopsis, DCL1 generates 21-nucleotide (nt) miRNAs, whereas DCL2, DCL3, and DCL4 produce siRNAs that are typically 22, 24, and 21 nt in length, respectively. These small RNAs program Argonaute (AGO)-containing effectors known as the RNA-induced silencing complex (RISC) that mediate sequence-specific translational inhibition, transcript cleavage, DNA methylation, or some combination of these, depending on the small RNAs and AGO proteins that they contain.
Dicer proteins possess RNase III domains and dsRNA binding motifs (dsRBMs), with the latter mediating sequence-nonspecific dsRNA binding as well as protein-protein interactions (10–12). An important group of Dicer-interacting factors includes proteins related to Caenorhabditis elegans RDE-4 (13). These proteins also have dsRBMs but lack an obvious catalytic domain. Some, for example, Drosophila R2D2 and R3D1/Loquacious, cooperate with specific Dicers in small RNA biogenesis and/or RISC loading (14, 15). Related proteins include TRBP and PACT in mammals, and in plants they comprise a family known simply as dsRNA binding proteins (DRBs). The DRB family in Arabidopsis has five members. The best studied is DRB1/HYL1, which interacts with DCL1, is required for efficient miRNA accumulation, and is involved in selecting the guide strand loaded into RISC (12, 16–19). DRB2 aids in processing specific miRNAs in the shoot apical meristem, suggesting considerable complexity in this developmentally critical tissue (20). DRB3 and DRB5 are dispensable for miRNA processing but have recently been shown to assist in silencing transcripts targeted by DRB2-associated miRNAs (21). DRB4 protein physically and functionally interacts with DCL4 in trans-acting siRNA (ta-siRNA) biogenesis and in siRNA-mediated defense against RNA viruses (18, 22–24). In addition, the Cauliflower mosaic virus (CaMV) P6 silencing suppressor interacts with DRB4, suggesting that a DRB4-DCL4 complex targets dsRNA derived from DNA virus transcripts (25). Although it has been suspected that at least one of the DRB proteins also plays a role in the nuclear RdDM pathway, studies have so far proved inconclusive, most likely because of functional redundancy. Mutational analysis to date has shown that none of the DRB proteins are essential for maintaining repressive transposon methylation (18, 26). However, DRB2 and DRB4 have been shown to inhibit the accumulation of small RNAs produced by RNA polymerase IV, a component of the RdDM pathway (27). Thus, multiple DRB proteins may have direct or indirect roles in the establishment or maintenance of RdDM.
Geminiviruses have small (2.5- to 3.0-kb) genomes of circular, single-stranded DNA that replicate in infected-cell nuclei by a rolling-circle mechanism that utilizes double-stranded DNA replicative-form (dsDNA RF) intermediates (28–30). Viral genomes specify four to seven proteins, none of which have polymerase activity. Instead, geminiviruses rely on host machinery for replication and transcription, both of which occur on viral chromatin templates composed of dsDNA RF and cellular histones organized as typical nucleosomes. Viral mRNAs are subject to posttranscriptional gene silencing (PTGS), and as a counterdefensive measure, geminivirus proteins suppress this cytoplasmic aspect of silencing (31, 32). In addition, we have shown that RNA-directed methylation of viral chromatin leading to transcriptional gene silencing (TGS) is a potent antiviral defense which is suppressed by geminivirus- and geminivirus satellite-encoded proteins (33–36). Genetic analysis has revealed that similar pathways are used to methylate geminivirus chromatin and resident transposons, suggesting that geminiviruses could serve as sensitive probes for the identification and analysis of methylation pathway components and effectors (4, 33, 37). Here, by analyzing the responses of drb mutant plants to geminivirus infection, we demonstrate that DRB3 acts with DCL3 and AGO4 in methylation-mediated defense against DNA viruses. We also show that DRB3 physically interacts with these critical effector proteins.
MATERIALS AND METHODS
Arabidopsis mutants.
DCL mutants were obtained from J. C. Carrington (38), and all others were obtained from the Arabidopsis Biological Research Center at The Ohio State University. The following seed stocks were used: wild-type (WT) Col-0 (CS60000) Columbia ecotype, drb2 (SALK_012017 and CS849395/At2g28380), drb3 (SALK_022644/At3g26932), drb4 (SALK_000736/At3g62800) (22), drb5 (SALK_031307C/At5g41071), dcl2-1 (SALK_064627/At3g03300), dcl3-1 (SALK_005512/At3g43920), dcl4-2 (GABI_160G05/At5g20320), wild-type Ler-0 (CS20) Landsberg erecta ecotype, and ago 4-1 gl1-1 (CS6364/At2g27040) (39). All mutants were homozygous, and when necessary this was verified by genotyping. Plants were reared in growth rooms at 22°C with 12-h light/dark cycles.
Virus inoculation.
Agroinoculation of Arabidopsis plants with Cabbage leaf curl virus (CaLCuV) or Beet curly top virus (BCTV) was carried out as previously described (33). Plants were inoculated within 5 days of bolting. Bolts were cut where they emerged from the rosette, and inoculum was applied to the freshly cut stem, which was then punctured with an insect pin multiple times. CaLCuV symptoms were observed and plants harvested at 14 to 21 days postinoculation; BCTV symptoms were observed and plants harvested at 21 to 30 days postinoculation. Inflorescence tissue showing visible symptoms was harvested from Arabidopsis. For each sample, tissue was pooled from four infected plants. For BCTV recovery experiments, plants were agroinoculated with BCTV or BCTV L2−, and after the primary harvest, plants were allowed to continue growing under the same conditions. The BCTV L2-1 null mutant has been previously described (40). Observations were made about symptom development in secondary inflorescence tissue, which was then harvested as secondary tissue 14 to 21 days after the harvesting of primary tissue.
BiFC.
Interactions between DCL and DRB proteins in plants were tested using bimolecular fluorescence complementation (BiFC) (41). The construction of BiFC expression vectors using enhanced yellow fluorescent protein (YFP) has been described previously (42). cDNA clones for DRB3 (U66272, 1,080 bp) and DRB4 (C104842, 1,099 bp) were obtained from the Arabidopsis Biological Resource Center. DCL3 and DCL4 cDNAs were obtained from J. C. Carrington. PCR primers were used to amplify these, with the introduction of a PacI site at the 5′ end and an AscI site at the 3′ end. The PCR products were subsequently digested with PacI and AscI and ligated into similarly digested BiFC vectors p2YN, p2YC, pYC, and pYN.
Cloned plasmids were transformed into Agrobacterium tumefaciens strain C58C1, and cultures were used to infiltrate 3- to 4-week-old Nicotiana benthamiana plants as previously described (43). Briefly, agrobacterium cells grown in liquid culture (optical density at 600 nm [OD600] = 1) were sedimented and incubated for 3 h in infiltration medium containing morpholineethanesulfonic acid (MES), acetosyringone, and MgCl2. Cultures containing p2YN- and p2YC-based plasmids were mixed 1:1 immediately prior to infiltration of young leaves using a blunt syringe. Red fluorescent protein (RFP)-histone 2B and RFP-fibrillarin were used as markers for the nucleus and nucleolus, respectively (44). Leaf tissue was analyzed by microscopy at approximately 36 h postinfiltration using a Nikon PCM 2000 confocal laser scanning microscope equipped with argon and green helium neon lasers with excitation wavelengths of 488 nm and 544 nm, respectively. To record YFP fluorescence, a band-pass emission filter (EM515/30HQ) with a 450- to 490-nm excitation wavelength and 515-nm emission wavelength was used. To record RFP fluorescence, a 565-nm long-pass filter (E565LP) was employed. Images were captured using Simple PCI software and compiled with Adobe Photoshop.
Co-IP and immunoblot analysis.
Immunoprecipitation (IP) experiments were performed to verify that DRB3 can physically associate with DCL3 and AGO4. Adenosine kinase (ADK) antibody was a negative control (45). Total protein extracts were prepared from leaves of transgenic dcl3 mutant Arabidopsis seedlings expressing a DCL3-FLAG fusion protein (a gift from Craig Pikaard) (46), N. benthamiana plants transiently expressing DRB3 fused with double-hemagglutinin, 6-histidine tags (HA2HIS6-DRB3), or N. benthamiana coexpressing HA2HIS6-DRB3 and FLAG-AGO4. The DRB3 and AGO4 proteins were expressed from the Tobacco mosaic virus-based TRBO vector as described previously (47). Immunoprecipitation was performed with monoclonal FLAG antibody (Sigma-Aldrich, F3165). To assess DRB3-DCL3 interaction, total protein extract (2.0 g) from leaves of Arabidopsis plants expressing DCL3-FLAG was mixed with 0.2 g extract from N. benthamiana leaves expressing HA2HIS6-DRB3. To verify DRB3-AGO4 interaction, total protein extract from 2 g of N. benthamiana leaves coexpressing HA2HIS6-DRB3 and FLAG-AGO4 was used. FLAG or ADK antibodies (10 μg) were incubated with protein G-agarose (Millipore) and protein extracts for 2 h at 4°C.
Proteins in immune complexes were fractionated by 10% SDS–PAGE (acrylamide/bisacrylamide ratio, 49:1) and analyzed by immunoblotting using 1:1,000 dilutions of FLAG antibody, ADK antibody, or monoclonal HA-peroxidase antibody (Sigma-Aldrich, H6533). Secondary antibodies included 1:1,000 dilutions of horseradish peroxidase (HRP)-linked anti-mouse IgG (Sigma-Aldrich, A5906) for anti-FLAG or HRP-linked anti-rabbit IgG (Sigma-Aldrich, A1949) for anti-ADK. Enhanced detection was performed using SuperSignal West Pico (for DRB3 and AGO4) or Femto (for DCL3) chemiluminescent substrate (Pierce).
Bisulfite sequencing.
Bisulfite sequencing was performed as described previously (33, 48). DNA isolated from infected plant tissue was linearized overnight using appropriate restriction enzymes (SacI for BCTV and XmnI for CaLCuV). Proteinase K digestion was subsequently carried out overnight, followed by bisulfite conversion using CT conversion reagent (EZ-DNA Methylation Gold; Zymo Research). Primers were designed against converted template, and the intergenic region (IR) of the viral genome was amplified by PCR. The PCR product was purified using Promega Wizard columns and TA cloned, and individual clones were sequenced at The Ohio State University Plant Microbe Genomics Facility. Conversion controls are discussed in Results. The forward and reverse primers used to amplify CaLCuV and BCTV IRs following bisulfite conversion are available upon request. Bisulfite reactions were carried out in a thermocycler. Data were analyzed and dot plots prepared using Kismeth (49).
Small RNA analysis.
RNA was isolated from 0.2 g of symptomatic floral tissue using Ribozol (Amresco) or TRIzol (Invitrogen) reagent. Each sample was pooled from 3 or 4 plants. Small RNAs were analyzed as described previously (24). Total RNA (5 to 8 μg) was loaded on a 16% denaturing polyacrylamide gel containing 8 M urea in 0.1× Tris-borate-EDTA (TBE). The gel was run until 10 min after the bromophenol blue dye ran off the gel. The gel was electroblotted onto a Hybond nylon membrane in 0.5× TBE at 4°C and 40 V for 1 h. RNA was cross-linked to the membrane using a Stratagene UV cross-linker at 1,700 kJ. The membrane was prehybridized in Ambion Ultrahyb oligonucleotide for 1 h at 40 to 42°C. A mixture of oligonucleotides (20 μM) was used as the probe. Oligonucleotides were labeled with Fermentas T4 polynucleotide kinase at 37°C for 30 to 60 min. Labeled probes were denatured, and hybridization was carried out overnight at 40 to 42°C. The membrane was washed 3 times for 20 min each with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% SDS at the same temperature. Ethidium bromide-stained rRNA was used as a loading control. Oligonucleotides used as size markers are available upon request, as are sense and antisense intergenic region and coat protein (CP) oligonucleotides that were used as hybridization probes.
RESULTS
Arabidopsis drb3 mutants are hypersusceptible to geminivirus infection.
Methylation-deficient Arabidopsis mutants exhibit hypersensitivity to geminiviruses that correlates with reduced cytosine methylation of the viral genome (33). To assess whether Arabidopsis DRB proteins might be involved in methylation-mediated defense, drb2, drb3, drb4, and drb5 mutant plants were challenged by geminivirus infection. DRB1 was not included in this study because it is known to participate exclusively in the miRNA pathway. Initial experiments were performed with Cabbage leaf curl virus (CaLCuV) (genus Begomovirus) and Beet curly top virus (BCTV) (genus Curtovirus) to evaluate responses to distinct types of geminiviruses. To permit observation of the full range of disease, inoculum doses that elicit mild symptoms in wild-type plants were used.
Geminivirus-infected drb2, drb4, and drb5 mutants showed mild to moderate disease enhancement relative to wild-type plants. Disease ratings with these mutants ranged from 1 to 2, where 1 indicates leaf curling and floral deformation typically seen with wild-type plants and 4 indicates very severe curling and deformation with extensive stunting (33). In contrast, disease symptoms in drb3 plants inoculated with either CaLCuV or BCTV were greatly enhanced and accompanied by considerable stunting (disease rating of 3 or 4) (Fig. 1A and B). Bisulfite sequencing was carried out to assess the methylation status of the viral intergenic region (IR). The IR (∼300 bp) contains the origin of replication flanked by divergent promoters, and in CaLCuV it includes 79 cytosines in different contexts, with 19 CG, 13 CNG, and 47 CHH sites (where H is any residue other than G). Following bisulfite treatment, the viral strand was amplified by PCR and 12 clones were sequenced per treatment.
FIG 1.
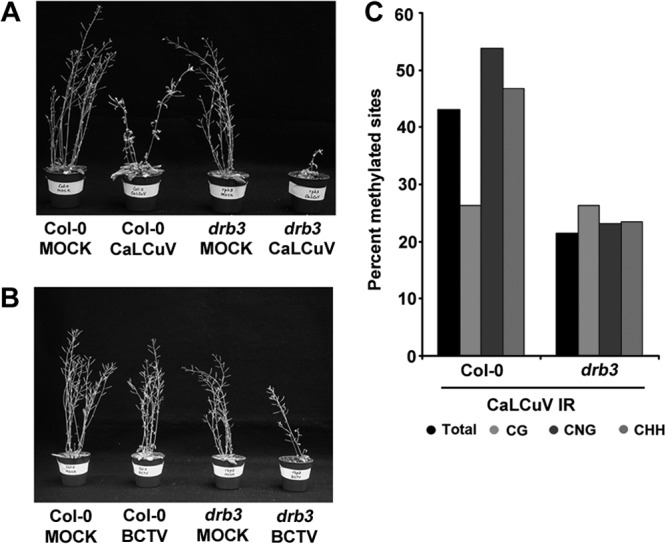
Arabidopsis drb3 mutants show enhanced susceptibility to geminivirus infection. (A) Photographs illustrate CaLCuV disease symptoms in wild-type (Col-0 ecotype) and drb3 mutant plants at 14 days postinoculation. (B) BCTV symptoms at 21 days postinoculation. BCTV has an inherently longer latent period than CaLCuV. (C) Histograms show the percentages of cytosines methylated in CaLCuV IR DNA isolated from wild-type and drb3 plants, as determined by bisulfite sequencing.
Bisulfite sequencing revealed that enhanced disease was accompanied by a substantial reduction in methylation. The proportion of methylated cytosine residues in the IR of drb3 mutants (∼20%) was considerably less than was observed in viral genomes obtained from wild-type plants (>40%, Col-0 ecotype) (Fig. 1C). We concluded that the phenotype of geminivirus-infected drb3 mutant plants was consistent with a defect in the methylation pathway.
Arabidopsis dcl3 and drb3 mutants are unable to recover from geminivirus infection.
Perhaps the most compelling argument for methylation as an antiviral defense comes from studies that have associated the methylation pathway with host recovery from geminivirus disease. Recovery occurs when tissues arising after the establishment of a systemic infection exhibit symptom remission. Recovery additionally is characterized by reduced viral DNA levels, as documented previously for several geminivirus-host combinations, including BCTV in N. benthamiana and Arabidopsis (33, 36, 40, 50, 51). Nicotiana benthamiana and Arabidopsis plants rarely recover from infection with wild-type BCTV but nearly always recover from infection with BCTV L2− mutant virus, which lacks the L2 (also known as C2) pathogenicity factor that suppresses both PTGS and TGS and nonspecifically inhibits methylation (33, 34, 40, 43, 45). Viral replication and transcription are inhibited by cytosine methylation (52, 53), and the small amount of geminivirus DNA found in recovered tissue is hypermethylated in the IR (33, 50, 51). In addition, we showed that ago4 plants are unable to recover from BCTV L2− infection and cannot hypermethylate the IR, confirming that the RdDM pathway is required for recovery (33). Based on these observations, we reasoned that an analysis of mutant plants for recovery might provide a sensitive and definitive means of identifying methylation pathway components.
To test this hypothesis, dcl2, dcl3, and dcl4 plants were examined. Considerable functional redundancy exists among Arabidopsis DCL enzymes, although DCL3 and the 24-nt siRNAs it generates are clearly associated with chromatin methylation (38, 54, 55). However, mild to moderate symptom enhancement was observed following BCTV or CaLCuV infection of Arabidopsis dcl2, dcl3, and dcl4 plants in earlier studies, precluding unequivocal identification of an individual DCL protein as the key player in methylation-mediated defense (33, 56). Similar results were again obtained following inoculation of these dcl mutants with BCTV L2−. However, after the establishment of systemic infections and removal of primary infected tissue, only dcl3 mutant plants proved unable to recover from infection. Severe symptoms appeared in all new axillary secondary shoots of all 32 dcl3 plants tested (Fig. 2). In contrast, symptom remission was observed in new shoots of wild-type, dcl2, and dcl4 plants (32 each). Thus, a host recovery assay could discriminate between functionally redundant DCL activities and identify DCL3 as the enzyme most associated with methylation-mediated defense.
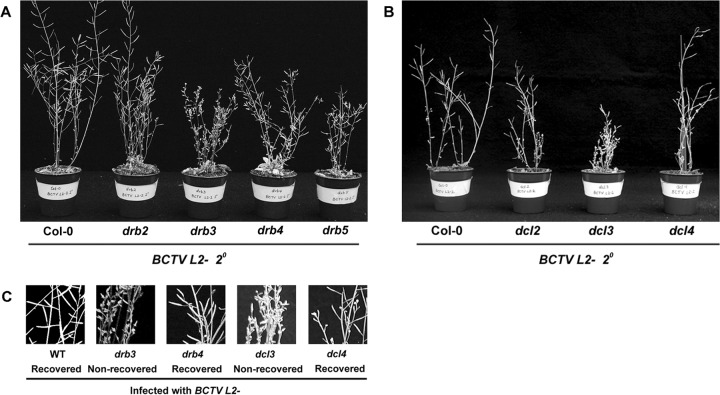
FIG 2.
Arabidopsis drb3 and dcl3 mutants do not recover from infection with BCTV L2− mutant virus. (A and B) The photographs show secondary tissue of wild-type (Col-0), drb2, drb3, drb4, and drb5 plants, as well as dcl2, dcl3, and dcl4 plants, infected with BCTV L2− virus. Note severe disease symptoms in the drb3 and dcl3 mutants and recovery (near absence of symptoms) in the wild type and all other mutants. Although deformation of floral tissue was not observed in secondary tissue of infected drb4 and drb5 mutants, plants were occasionally stunted. Examples of stunted drb4 and drb5 plants are shown in this figure. (C) Close-up views of infected shoots from drb3, drb4, dcl3, and dcl4 plants.
To confirm a role for DRB3 in methylation, a panel of drb mutant plants was inoculated with BCTV L2−, and, as observed with wild-type BCTV, symptoms were much more severe in drb3 mutants. As expected, new secondary shoots of wild-type plants infected with BCTV L2− recovered from infection and showed little evidence of disease, as did nearly all new shoots of the drb2, drb4, and drb5 mutants (32 plants each). However, while new tissues of drb4 and drb5 plants never displayed floral deformation, a small minority of the plants were somewhat stunted. Examples of these outliers are shown in Fig. 2. In contrast, drb3 mutants did not recover, and severe symptoms appeared in all new shoots of the 32 drb3 plants inoculated with BCTV L2− (Fig. 2). Together with previous studies of ago4 mutants (33), these experiments confirm that RdDM conditioned by DCL3 and AGO4 is required for host recovery from geminivirus infection, and they provide strong genetic evidence linking DRB3 to this pathway.
Arabidopsis dcl3 and drb3 mutants cannot hypermethylate the viral genome.
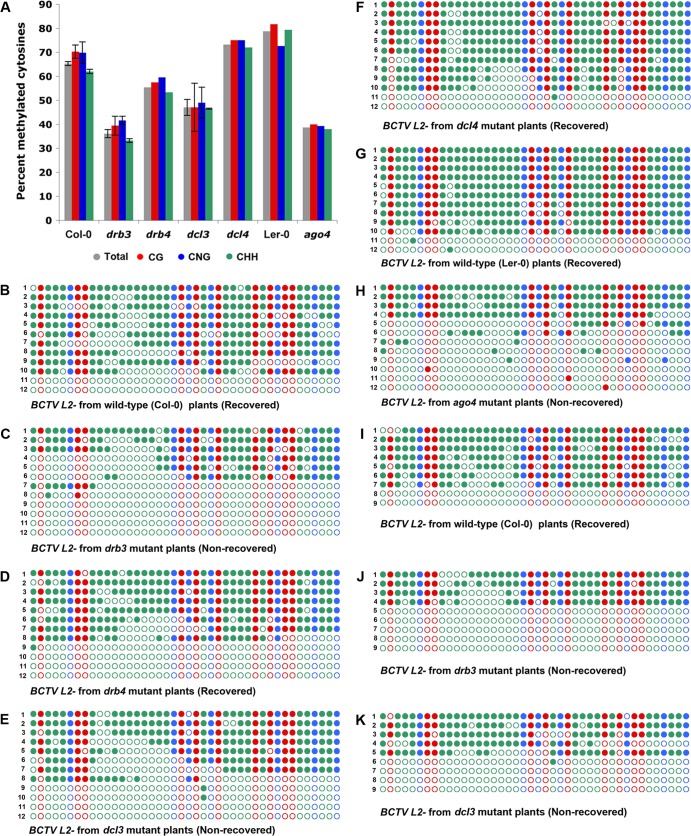
We previously showed that the relatively small amount of BCTV L2− DNA in recovered wild-type plants is hypermethylated, while the same virus from comparable but nonrecovered tissue of ago4 mutants is not (33). Thus, we compared the methylation status of BCTV L2− DNA obtained from recovered tissue with that from nonrecovered tissue of drb3, dcl3, and ago4 mutants. Mutant drb4 and dcl4 plants were included in this analysis because a DRB4-DCL4 complex is involved in PTGS-mediated defense against RNA viruses and DNA virus transcripts (24, 25, 56–58). Bisulfite sequencing was carried out as before, except that primers designed to amplify the viral strand of the BCTV IR were employed. The BCTV IR contains 42 cytosines in the following contexts: 10 CG, 7 CNG, and 25 CHH. PCR products were cloned and sequenced. It should be noted that, to preclude common artifacts of bisulfite sequencing, rigorous conversion controls are routinely included in all bisulfite experiments. Here, as an internal control, a bacterial plasmid containing CaLCuV DNA was added to BCTV-infected sample extracts prior to treatment with bisulfite reagent. Alternatively, as a parallel control, a plasmid containing BCTV DNA was added to an uninfected plant DNA extract. Data were accepted only when control DNA conversion was complete (12 clones analyzed per treatment). In some cases, plasmid DNA was methylated in vitro with CG methylase before being added to plant extracts. In these cases, data were accepted only when conversion was complete at non-CG sites and all CG sites remained unconverted. Bisulfite sequencing data for all plants examined are illustrated graphically in Fig. 3A. Cytosine methylation profiles representing individual BCTV L2− IR clones obtained from wild-type (Col-0), drb3, drb4, dcl3, and dcl4 plants, as well as wild-type (Ler-0) and ago4 mutant plants, are shown in Fig. 3B to K. Unlike the drb and dcl mutations, the ago4 mutation is in the Ler-0 ecotype background.
FIG 3.
Arabidopsis drb3 and dcl3 mutants fail to hypermethylate the viral genome. Methylation of BCTV L2− IR DNA was assessed by bisulfite sequencing. (A) Histograms indicate the percentages of cytosine residues methylated in different sequence contexts in viral DNA obtained from secondary tissues of wild-type (Col-0 or Ler-0) or mutant plants. The drb and dcl mutants are in Col-0, whereas ago4 is in the Ler-0 ecotype background. Bars indicate the mean ± standard error (SE). Also shown are cytosine methylation profiles of BCTV L2− DNA obtained from recovered wild-type Col-0 (B), nonrecovered drb3 (C), recovered drb4 (D), nonrecovered dcl3 (E), recovered dcl4 (F), recovered wild-type Ler-0 (G), and nonrecovered ago4 (H) plants. A second, independent experiment included recovered wild-type Col-0 (I), nonrecovered drb3 (J), and nonrecovered dcl3 (K) plants. The dots represent all cytosines in the IR and are color coded according to sequence context (red, CG; blue, CNG; green, CHH). Filled circles indicate methylation, and each line represents the sequence of an individual clone, arranged from most to least methylated. Significant reductions in methylation were noted for drb3 (P = 0.013) compared to wild-type controls, as determined by the Mann-Whitney rank sum test.
The majority of genomes from recovered tissues of wild-type plants were hypermethylated in all sequence contexts, with ∼63% (Col-0) to ∼77% (Ler-0) of total cytosines methylated (Fig. 3A to H). In contrast, methylation levels were substantially reduced in viral genomes obtained from drb3, dcl3, and ago4 mutants. Specifically, considerably less methylation was observed in nonrecovered drb3 plants than in recovered drb4 plants (34% and 54%, respectively) and in nonrecovered dcl3 plants than in recovered dcl4 plants (48% and 70%, respectively). The greatest reduction relative to the comparable wild type (Ler-0) was noted in genomes from ago4 mutant plants (38% compared to 77%). In a second, independent experiment involving wild-type, drb3, and dcl3 plants, similar results were obtained. In this case, the proportions of cytosines methylated were 62%, 35%, and 43%, respectively (Fig. 3I to K). Given that similar outcomes were observed with drb3, dcl3, and ago4 plants in independent experiments reported here and in previous studies, it is highly unlikely that our results were significantly impacted by selection of sibling clones for bisulfite sequencing. These results confirm that geminivirus genomes obtained from recovered tissues (wild type, drb4, and dcl4) are more densely methylated than those from nonrecovered drb3, dcl3, ago4 plants, and they further associate DRB3 with the antiviral RdDM pathway that involves DCL3 and AGO4.
Both active and repressed viral genomes are present in infected plants.
Histone H3 associated with geminivirus DNA carries modifications characteristic of both active (acetylated H3 [H3Ac]) and repressed (H3K9me2) chromatin (33), and cytosine methylation profiles reveal mixtures of mostly hypermethylated (likely repressed) or mostly hypomethylated (likely active) genomes (Fig. 3). Together, these observations suggest that populations of active and repressed viral genomes coexist in infected plants and that the major effect of specific host or viral mutations that lead to hypersusceptibility or recovery is to alter their relative proportions.
Chromatin immunoprecipitation followed by bisulfite sequencing (ChIP-BS) was employed to address this issue. Beginning with nonrecovered tissue from a BCTV L2−-infected mutant plant (input), ChIP-BS was performed with antibody specific for H3Ac (acetylated at lysine 9 or 14) or H3K9me2, and precipitated DNA was treated with bisulfite reagent. Following PCR amplification with IR primers, clones were obtained and sequenced. We observed that H3Ac precipitation led to a moderate enrichment in hypomethylated genomes compared to input DNA (∼40% compared to ∼50% of cytosines methylated). In contrast, all of the genomes associated with H3K9me2, a hallmark of repressed chromatin, were hypermethylated (∼90% of cytosines methylated) (Fig. 4A and B). Similar results were obtained in a second, independent experiment carried out using recovered tissue from a plant infected with BCTV L2−, where the viral DNA (input) was already hypermethylated. Precipitation with H3Ac antibody again led to a moderate enrichment of hypomethylated genomes relative to input DNA (∼55% compared to ∼70% of cytosines methylated), and all genomes associated with H3K9me2 were hypermethylated (∼90% of cytosines methylated) (Fig. 4C and D). These findings support the view that populations of active and repressed genomes are present in infected plants and indicate that even during a productive, symptomatic infection, a significant fraction of viral genomes are silenced by both cytosine and H3K9 methylation.
FIG 4.
Hypermethylated viral genomes are associated with H3K9me2. ChIP-BS was performed with extracts from BCTV L2−-infected, nonrecovered primary tissue using antibodies to H3Ac or H3K9me2. Input (extract without ChIP) and ChIP DNAs were bisulfite treated, and PCR primers spanning the viral IR were used to amplify associated DNA. Products were cloned and sequenced. (A) Histograms indicate the percentages of cytosine residues methylated in different sequence contexts. (B) Cytosine methylation profiles. (C and D) In a second, independent experiment, ChIP-BS was performed with extracts from BCTV L2− infected, recovered secondary tissue, where the input DNA was already hypermethylated. The dots represent all cytosines in the IR and are color coded according to sequence context (red, CG; blue, CNG; green, CHH). Filled circles indicate methylation, and each line represents the sequence of an individual clone, arranged from most to least methylated. The difference between the nonrecovered input and H3K9me2 fractionated samples was significant (P = 0.007), as determined by the Mann-Whitney rank sum test.
DRB3 interacts with DCL3 and AGO4.
The similar recovery phenotypes observed with ago4, dcl3, and drb3 mutant plants prompted us to ask whether these proteins physically interact. We first employed bimolecular fluorescence complementation (BiFC) to examine in vivo interactions between these proteins, using DCL4 and DRB4 as a positive interaction control. BiFC involves fusion of potentially interacting proteins with the N- or C-terminal portions of yellow fluorescent protein (YFP) and subsequent coexpression (41). Protein association reconstitutes YFP, producing a fluorescent signal that at once reveals the interaction and where it occurs in the cell. In these studies, N. benthamiana leaves were coinfiltrated with mixtures of Agrobacterium tumefaciens cells harboring constructs that express the fusion proteins (42). Previous studies with this system have shown that expression of Arabidopsis proteins in N. benthamiana yields biologically relevant results (36, 42, 59). Test proteins were expressed as both N- and C-terminal fusions with both the N- and C-terminal portions of YFP. Proteins were judged to interact when signal was observed in multiple combinations and not to interact when signal was absent in all combinations.
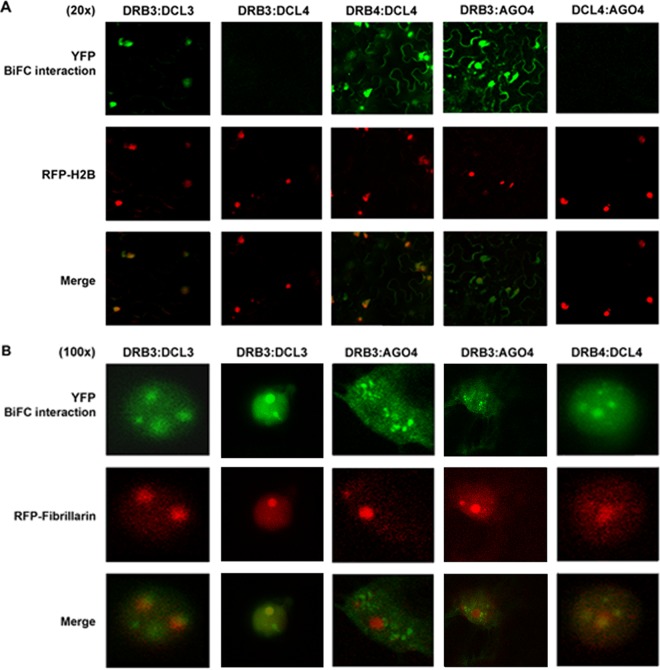
In agreement with genetic evidence linking DRB3 to the methylation pathway, we found that DRB3 and DCL3 interact in the nucleus (Fig. 5A). The DRB3-DCL3 signal was observed throughout and was especially intense in subnuclear bodies, some but not all of which were associated with nucleoli (Fig. 5B). This is consistent with previous localization of DCL3, AGO4, and siRNA in Cajal bodies, ribonucleoprotein processing centers that sometimes associate with nucleoli (46). In contrast, no evidence of complex formation was observed when DRB3 and DCL4 fusion proteins were coexpressed. Thus, our data show that DRB3 specifically interacts with the RdDM-associated DCL3 and not with PTGS-associated DCL4. As a positive interaction control, we confirmed that DRB4 interacts with DCL4 (12, 23). DRB4-DCL4 complexes were similarly observed in nucleolus-associated bodies, although they were also present in the cytoplasm (Fig. 5A and B).
FIG 5.
DRB3 interacts with DCL3 and AGO4 in distinct subnuclear bodies. BiFC analysis of the indicated DCL, DRB, and AGO proteins in N. benthamiana epidermal cells was performed. Constructs expressing proteins fused to the N- or C-terminal portion of YFP were delivered by agroinfiltration to N. benthamiana leaves. Cells were photographed at 36 h postinfiltration using a confocal laser scanning microscope. RFP-histone 2B (RFP-H2B) and RFP-fibrillarin were used as markers for the nucleus and nucleolus, respectively. Protein combinations are indicated above each photograph. The photographs in panel A show lower-magnification views (×20), while panel B shows high-magnification views of nuclei (×100).
AGO4 has been reported to localize in two distinct subnuclear bodies, namely, Cajal bodies and smaller AB bodies, which also contain RNA polymerase V and DRM2 (46, 60, 61). We observed that DRB3 interacts with AGO4 throughout the nucleus, although some signal was also apparent in the cytoplasm (Fig. 5A). Nuclear complexes were concentrated in small punctate spots that were distinct in appearance from the larger bodies containing DRB3-DCL3 and did not associate with nucleoli (Fig. 5B).
Coimmunoprecipitation (co-IP) experiments were carried out to confirm the interactions. Unfortunately, despite considerable effort, we were unable to express DCL3 in yeast, plant, or Escherichia coli cells using standard methods. Thus, dcl3 mutant, transgenic Arabidopsis plants expressing a complementing DCL3-FLAG transgene were employed (46). Arabidopsis DRB3 was expressed as a double-hemagglutinin peptide, 6-histidine fusion (HA2His6-DRB3), and AGO4 as a FLAG fusion (FLAG-AGO4), in N. benthamiana plants using a Tobacco mosaic virus-based vector (47). When protein extracts containing FLAG-DCL3 and HA2His6-DRB3 were mixed and incubated with FLAG antibody, both DCL3 and DRB3 were immunoprecipitated (Fig. 6A). Neither protein was precipitated using a control antibody raised against adenosine kinase (ADK). Similarly, when extracts from N. benthamiana plants coexpressing FLAG-AGO4 and HA2His6-DRB3 were incubated with FLAG antibody, both AGO4 and DRB3 were immunoprecipitated (Fig. 6B). Together, these BiFC and co-IP studies demonstrate physical associations between DRB3-DCL3 and DRB3-AGO4 that are consistent with functional cooperation in methylation-mediated antiviral defense.
FIG 6.
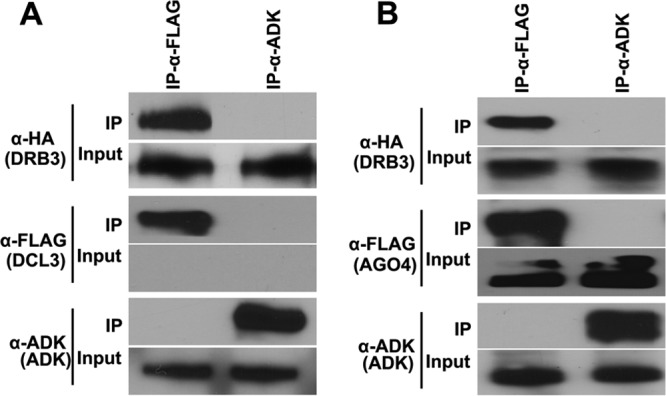
DRB3 coimmunoprecipitates with DCL3 and AGO4. Immunoprecipitation (IP) was performed with FLAG antibody (α-FLAG) or ADK antibody (α-ADK, negative control), and proteins in immune complexes were detected by Western blotting using HA antibody (α-HA), α-FLAG, or α-ADK. (A) Extracts from plants expressing DCL3-FLAG or HA2His6-DRB3 were mixed at ∼10:1 to compensate for extremely low levels of DCL3-FLAG expression (below the level of detection in input samples). (B) Extracts were obtained from plants coexpressing FLAG-AGO4 and HA2His6-DRB3.
DRB3 is not required for the biogenesis of 24-nt siRNAs by DCL3.
While some dsRBM proteins do not significantly impact small RNA levels, others facilitate the biogenesis of small RNA species by the Dicers with which they associate. We asked whether DRB3 is necessary for the accumulation of 24-nt siRNA generated by DCL3 in response to BCTV. While it was previously shown that levels of 24-nt siRNA derived from an RNA virus were not diminished in drb3 plants (18), virus-specific siRNAs of this size class are considerably more abundant in geminivirus-infected plants (56, 62).
RNA was isolated from wild-type, drb3, and dcl3 plants inoculated with BCTV, and infected drb4, dcl4, and ago4 plants were included in this study for comparison. RNA samples were fractionated on polyacrylamide gels and probed with a mixture of 32P-labeled oligonucleotides specific for the BCTV IR and the coat protein (CP)-coding region. Previous experiments using separate IR or CP oligonucleotide probes have shown that both regions spawn abundant BCTV-derived siRNAs of all size classes (data not shown).
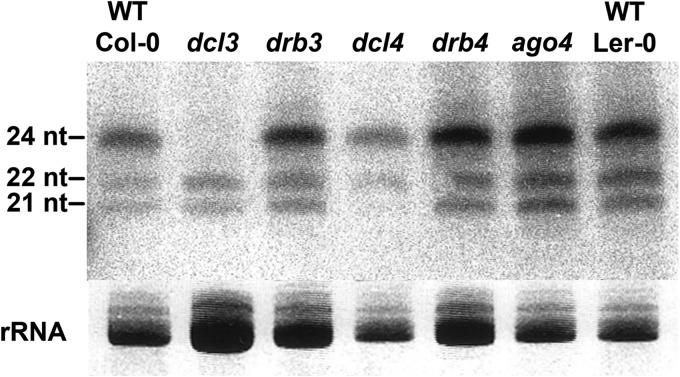
As expected, dcl3 mutants lacked 24-nt siRNAs corresponding to the BCTV genome, and dcl4 mutants showed much reduced 21-nt siRNA levels (Fig. 7). However, little change was evident in the type and abundance of small RNA species present in the drb3, drb4, and ago4 mutant plants compared to corresponding wild-type plants (Col-0 or Ler-0). We concluded that DRB3 and DRB4 are not required for the synthesis or accumulation of BCTV-derived siRNAs.
FIG 7.
DRB3 is not required for biogenesis of geminivirus-derived siRNAs. RNA blot hybridization shows the accumulation of virus-specific siRNAs in BCTV-infected wild-type (Col-0 and Ler-0) and mutant (drb3, drb4, dcl3, dcl4, and ago4) plants. Ethidium bromide-stained rRNA served as a loading control. The positions of oligonucleotide size markers are indicated at the left.
DISCUSSION
Nuclear RdDM that involves DCL3 and AGO4 limits geminivirus disease, and the goal of this study was to determine whether an Arabidopsis DRB protein participates in this epigenetic defense. By taking advantage of the extreme sensitivity of methylation-deficient plants to geminivirus infection, we generated genetic evidence that strongly links DRB3 with this pathway. We found that drb3 plants show enhanced susceptibility to geminivirus infection that is accompanied by reduced levels of cytosine methylation in the viral IR. In addition, like dcl3 and ago4 mutants, drb3 plants fail to recover from infection with a virus lacking a suppressor protein that inhibits methylation (BCTV L2−) and are unable to carry out the IR hypermethylation that is typically observed in recovered wild-type plants. Significantly less methylation was observed in nonrecovered drb3 plants than in recovered wild-type plants (∼34 to 35% and 62 to 63% methylated cytosines, respectively).
Physical interaction is another indication of functional cooperation between DRB3, DCL3, and AGO4. We found that DRB3-DCL3 complexes preferentially accumulate in relatively large bodies that are occasionally associated with nucleoli, similar to Cajal bodies known to contain DCL3, AGO4, and siRNA (46). Surprisingly, DRB3-AGO4 complexes were not seen in these structures but were enriched in smaller, punctate spots reminiscent of AB bodies that contain AGO4, RNA polymerase V, and DRM2 (46, 60, 61). Why sites of DRB3-DCL3 and DRB3-AGO4 complex enrichment do not overlap is unclear, but this raises the intriguing possibility that DRB3 choreographs nuclear events by interacting with upstream and downstream pathway components in different locations. It will be of interest to determine, in future studies, whether virus infection alters the localization or interactions of these silencing pathway components. DRB3-DCL3 interaction was confirmed in coimmunoprecipitation experiments in which, for technical reasons, interacting proteins were separately expressed in different plants. Nevertheless, DRB3-DCL3 complexes were able to form after the plant extracts were mixed. DRB3-AGO4 interaction was verified by co-IP following coexpression of these proteins in N. benthamiana plants. An earlier study showed that a DRB3-YFP fusion protein was present in the cytoplasm following transient expression in N. benthamiana cells, although this study did not rule out the existence of a nuclear pool of DRB3 (20). Cytoplasmic localization is consistent with a downstream role for DRB3 in the miRNA pathway (21). Nuclear localization of DRB3-DCL3 and DRB3-AGO4 complexes is consistent with an additional role for DRB3 in the methylation pathway, and since BiFC will detect proteins only when they are in complex, a cytoplasmic pool of DRB3 would have escaped detection in our analysis.
While our experiments clearly show that drb3 mutant plants are methylation deficient from the perspective of geminiviruses, they differ from an earlier study which found that methylation levels of several cellular transposons were unaltered in drb3 plants or in any of the drb mutants, including multiple mutants (18). We suggest that this may be attributable in part to functional redundancy and further that the enormous demand imposed on the methylation pathway by geminivirus infection allowed us to detect reduced viral DNA methylation in drb3 plants which could not be observed at endogenous loci. In support of this view, the results of our ChIP-BS experiments indicate that a substantial fraction of the viral genomes in infected plants (which can approach 0.5 × 106 copies per cell) are repressed by dense cytosine methylation and by association with H3K9me2 (Fig. 4). Thus, we argue that DRB3 is the primary DRB protein involved in the methylation-mediated antiviral defense. While paralogues of DRB3, DCL3, and AGO4 are capable of directing viral genome methylation in the absence of these enzymes, the functionally redundant activities are clearly not sufficient to generate the very high methylation levels necessary to support recovery. It should also be mentioned that while DRB3 expression can be detected in all tissues, levels are particularly high in apical and axillary shoots where recovery is initiated (20, 63). However, we cannot rule out the possibility that, rather than redundant DRB, DCL, or AGO activities, noncanonical pathways are responsible for geminivirus genome methylation observed in nonrecovered drb3, dcl3, and ago4 mutant plants.
Maintenance of CG and CNG methylation by the methyltransferases MET1 and CMT3, respectively, is another factor that might obscure a role for DRB3 at transposons and other endogenous sequences. Nonsymmetrical CHH methylation is not robustly recognized by a maintenance methyltransferase, and although a CMT2-dependent pathway was recently shown to be involved in asymmetric methylation of heterochromatin, CHH methylation in Arabidopsis is at most 20% at specific loci and averages less than 2% genome wide (64–66). Geminivirus dsDNA replicative forms, on the other hand, must be methylated de novo in infected cells, and maintenance thorough mitosis is not required. Evidence of this is the large proportion of CHH methylation that we routinely observe on the BCTV IR, which can reach 60 to 80% in wild-type Col-0 and Ler-0 plants infected with a mutant virus that cannot suppress methylation (BCTV L2−). Similar levels of CHH methylation are seen when silencing is reestablished on transposons that were recently transcriptionally activated, reinforcing the notion that dense CHH methylation is characteristic of de novo methylation (67). Thus, it is possible that DRB3 is primarily involved in the establishment of methylation, and if so, the effects of its absence would be obvious on viral genomes but difficult to detect at resident transposons.
It bears repeating that rigorous conversion controls are employed in all bisulfite experiments (see Results), and we are confident that the pattern of mostly methylated or mostly unmethylated clones that we routinely observe is accurate. However, it is worth considering that geminivirus DNAs exist in multiple forms in infected cells: (i) circular single-stranded DNA (ssDNA), which cannot be methylated; (ii) circular dsDNA replicative forms, which are templates for replication and transcription and may or may not be methylated; and (iii) linear dsDNA forms of heterogeneous length, some of which may be products of recombination-dependent replication, that also may or may not be methylated (68, 69). By Southern blotting, we have not observed reproducible alterations in the ratio of circular ssDNA to dsDNA in any of the methylation-deficient mutants we have examined. Thus, reductions in viral genome methylation that characteristically occur in known methylation-deficient mutants cannot be attributed to changes in viral ssDNA and dsDNA levels. In addition, previous studies of Tomato yellow leaf curl China virus and BCTV have shown that during primary infections, most viral genome methylation is found at sequences encompassing or adjacent to viral promoters, including the IR (reference 36 and unpublished results). Similar results have been obtained with other geminiviruses (51). This methylation could exist either on circular dsDNA forms or on heterogenous linear DNA or on both. It has been reported that the bulk of methylated geminivirus DNA exists as heterogeneous linear dsDNA forms which presumably correspond to all regions of the viral genome (70). Thus, it is possible that linear viral DNA is preferentially methylated and/or that methylated and repressed circular dsDNA forms accumulate double-strand breaks. Circular dsDNA forms in cells where the virus is not suppressed will be highly amplified by rolling-circle replication, which minimizes methylation. Our bisulfite sequencing protocol does not discriminate between viral DNAs of different conformations, and, of course, the level of methylation observed over any particular region or with any particular virus is to some extent biased by the primers used to amplify converted DNA. Nevertheless, we observe reproducible differences in the proportion of methylated to unmethylated viral IR clones obtained from wild-type plants or nonmethylation mutants (e.g., dcl4) compared with those obtained from mutant plants that are known to be methylation deficient (e.g., dcl3 and ago4). Further, we see the highest methylation levels in recovered wild-type plants using a virus (BCTV L2−) that lacks a suppressor protein known to inhibit methylation. Thus, our results accurately reflect relative methylation levels of aggregate viral DNA populations in infected cells.
Small RNA gel blot experiments showed that the absence of DRB3 does not significantly impact the biosynthesis or accumulation of geminivirus-derived siRNAs, including the 24-nt size class generated by DCL3. That methylation of viral genomes clearly occurs in dcl3 mutant plants suggests that small RNAs processed by other DCL enzymes can redundantly participate in the methylation pathway or that noncanonical mechanisms contribute to geminivirus genome methylation. In any case, the absence of an effect of DRB3 on small RNA profiles is not unprecedented, as some DRB proteins appear to have little or no role in small RNA biogenesis. For example, only a small decrease in 21-nt virus-specific siRNA levels was observed in drb4 plants infected with Turnip crinkle virus, an RNA virus (24). Further, although DRB3 was recently shown to participate in miRNA-mediated silencing of specific transcripts, it has no role in miRNA biogenesis (21). Thus, DRB3 appears to act downstream of both DCL1/DRB2-mediated miRNA processing and DCL3-mediated 24-nt siRNA processing.
Similar to drb3, the ago4 mutation also did not cause a loss or reduction of BCTV-specific siRNAs. This is at odds with a previous study that reported a reduction in 24-nt siRNAs originating from some endogenous loci in the absence of AGO4, and it was proposed that AGO4 binding might stabilize siRNAs (71). We suggest that our disparate results may be due to the extreme abundance of geminivirus-derived siRNAs in infected plants, which could saturate RISCs and obscure a stabilizing effect of siRNA binding.
In conclusion, we set out to determine if a particular DRB protein(s) contributes in a significant way to geminivirus genome methylation, and our results strongly implicate DRB3. Beginning with an unbiased screen of drb mutants, we demonstrated the following: (i) drb3 mutants uniquely show enhanced susceptibility to geminiviruses; (ii) drb3 mutants cannot recover from infection; (iii) drb3 mutants fail to hypermethylate the viral genome, which is essential for recovery; (iv) the response of drb3 mutants to geminivirus infection is essentially identical to that of dcl3 and ago4 mutant plants, which are defective for well-characterized components of the RdDM pathway; and (v) DRB3 physically associates with DRB3 and AGO4 in the nucleus. Thus, although participation of DRB3 in cellular DNA methylation was not addressed and remains open to debate, these studies provide unequivocal evidence that DRB3 plays an important role in the RdDM pathway that conditions an epigenetic defense against geminiviruses.
ACKNOWLEDGMENTS
We thank Debbie Parris, Keith Slotkin, Biao Ding, Erich Grotewold, and Iris Meier for helpful discussions. We are grateful to Feng Qu for advice on small RNA analysis, Jim Carrington for providing dcl mutants, Michael Goodin for RFP-histone 2B and RFP-fibrillarin, Greg Booton for advice on statistical analysis of bisulfite sequencing data, and Craig Pikaard for transgenic Arabidopsis expressing DCL3-FLAG. We also thank members of the Bisaro and Slotkin labs for advice and technical support.
This work was supported by NSF grants MCB-0743261 and MCB-1158262 to D.M.B. J.N.J. was supported by predoctoral fellowships from the OSU Center for RNA Biology and from the Pelotonia program of the OSU Comprehensive Cancer Center.
Footnotes
Published ahead of print 18 December 2013
REFERENCES
- 1.Baulcombe DC. 2004. RNA silencing in plants. Nature 431:356–363. 10.1038/nature02874 [DOI] [PubMed] [Google Scholar]
- 2.Brodersen P, Voinnet O. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22:268–280. 10.1016/j.tig.2006.03.003 [DOI] [PubMed] [Google Scholar]
- 3.Vaucheret H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20:759–771. 10.1101/gad.1410506 [DOI] [PubMed] [Google Scholar]
- 4.Henderson IR, Jacobsen SE. 2007. Epigenetic inheritance in plants. Nature 447:418–424. 10.1038/nature05917 [DOI] [PubMed] [Google Scholar]
- 5.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM. 2009. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21:367–376. 10.1016/j.ceb.2009.01.025 [DOI] [PubMed] [Google Scholar]
- 6.Law JA, Jacobsen SE. 2010. Establishing, maintaining, and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11:204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding S-W, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426. 10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mlotshwa S, Pruss GJ, Vance V. 2008. Small RNAs in viral infection and host defense. Trends Plant Sci. 13:375–382. 10.1016/j.tplants.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Ferrer V, Voinnet O. 2009. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60:485–510. 10.1146/annurev.arplant.043008.092111 [DOI] [PubMed] [Google Scholar]
- 10.Saunders LR, Barber GN. 2003. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 17:961–983. 10.1096/fj.02-0958rev [DOI] [PubMed] [Google Scholar]
- 11.Chang K-Y, Ramos A. 2005. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 272:2109–2117. 10.1111/j.1742-4658.2005.04652.x [DOI] [PubMed] [Google Scholar]
- 12.Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. 2005. Specific interacions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57:173–188. 10.1007/s11103-004-6853-5 [DOI] [PubMed] [Google Scholar]
- 13.Tabara H, Yigit E, Siomi H, Mello C. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109:861–871. 10.1016/S0092-8674(02)00793-6 [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Rand TA, Kalidas S, Du F, Kim H-E, Smith DP, Wang X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921–1925. 10.1126/science.1088710 [DOI] [PubMed] [Google Scholar]
- 15.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. 2005. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 19:1674–1679. 10.1101/gad.1334005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han M-H, Goud S, Song L, Federoff N. 2004. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U. S. A. 101:1093–1098. 10.1073/pnas.0307969100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasquez F, Gasciolli V, Crete P, Vaucheret H. 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14:348–351. 10.1016/j.cub.2004.01.035 [DOI] [PubMed] [Google Scholar]
- 18.Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM. 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 582:2753–2760. 10.1016/j.febslet.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 19.Eamens AL, Smith NA, Curtin S, Wang M-B, Waterhouse PM. 2009. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15:2219–2235. 10.1261/rna.1646909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eamens AL, Kim KW, Curtin SJ, Waterhouse PM. 2012. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS One 7:e35933. 10.1371/journal.pone.0035933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eamens AL, Wook KK, Waterhouse PM. 2012. DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal. Behav. 7:1224–1229. 10.4161/psb.21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. 2006. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16:927–932. 10.1016/j.cub.2006.03.035 [DOI] [PubMed] [Google Scholar]
- 23.Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. 2007. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol. Biol. 63:777–785. 10.1007/s11103-006-9125-8 [DOI] [PubMed] [Google Scholar]
- 24.Qu F, Ye X, Morris TJ. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105:14732–14737. 10.1073/pnas.0805760105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas G, Azevado J, Moissard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O. 2008. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 27:2102–2112. 10.1038/emboj.2008.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eamens AL, Curtin SJ, Waterhouse PM. 2011. The Arabidopsis thaliana double-stranded RNA binding (DRB) domain protein family, p 385–403 In Erdmann VA, Barciszewski J. (ed), Noncoding RNAs in plants. Springer, Berlin, Germany [Google Scholar]
- 27.Pelissier T, Clavel M, Chaparro C, Pouch-Pelissier M-N, Vaucharet H, Deragon J-M 2011. Double-stranded RNA binding proteins DRB2 and DRB4 have an antagonistic impact on polymerase IV-dependent siRNA levels in Arabidopsis. RNA 17:1502–1510. 10.1261/rna.2680711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43:361–394. 10.1146/annurev.phyto.43.040204.135939 [DOI] [PubMed] [Google Scholar]
- 29.Jeske H. 2009. Geminiviruses. Curr. Top. Immunol. Microbiol. 331:185–226 [DOI] [PubMed] [Google Scholar]
- 30.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11:777-788. 10.1038/nrmicro3117 [DOI] [PubMed] [Google Scholar]
- 31.Vanitharani R, Chellappan P, Fauquet CM. 2005. Geminiviruses and RNA silencing. Trends Plant Sci. 10:144–151. 10.1016/j.tplants.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 32.Bisaro DM. 2006. Silencing suppression by geminivirus proteins. Virology 344:158–168. 10.1016/j.virol.2005.09.041 [DOI] [PubMed] [Google Scholar]
- 33.Raja P, Sanville BC, Buchmann RC, Bisaro DM. 2008. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82:8997–9007. 10.1128/JVI.00719-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. 2009. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83:5005–5013. 10.1128/JVI.01771-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raja P, Wolf JN, Bisaro DM. 2010. RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochim. Biophys. Acta 1799:337–351. 10.1016/j.bbagrm.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, Bisaro DM, Zhou X. 2011. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7:e1002329. 10.1371/journal.ppat.1002329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lisch D. 2009. Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 60:43–66. 10.1146/annurev.arplant.59.032607.092744 [DOI] [PubMed] [Google Scholar]
- 38.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:e104. 10.1371/journal.pbio.0020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zilberman D, Cao X, Jacobsen SE. 2003. Argonaute4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716–719. 10.1126/science.1079695 [DOI] [PubMed] [Google Scholar]
- 40.Hormuzdi SG, Bisaro DM. 1995. Genetic analysis of beet curly top virus: examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206:1044–1054. 10.1006/viro.1995.1027 [DOI] [PubMed] [Google Scholar]
- 41.Hu C-D, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins using bimolecular fluorescence complementation. Mol. Cell 9:789–798. 10.1016/S1097-2765(02)00496-3 [DOI] [PubMed] [Google Scholar]
- 42.Yang X, Baliji S, Buchmann RC, Wang H, Lindbo JA, Sunter G, Bisaro DM. 2007. Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J. Virol. 81:11972–11981. 10.1128/JVI.00617-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM. 2005. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79:7410–7418. 10.1128/JVI.79.12.7410-7418.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakrabarty R, Banerjee R, Chung S-M, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M. 2007. pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20:740–750. 10.1094/MPMI-20-7-0740 [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Hao L, Shung C-Y, Sunter G, Bisaro DM. 2003. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020–3032. 10.1105/tpc.015180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. 2006. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126:79–92. 10.1016/j.cell.2006.05.031 [DOI] [PubMed] [Google Scholar]
- 47.Lindbo JA. 2007. TRBO: a high-efficiency tobacco mosiac virus RNA-based overexpression vector. Plant Physiol. 145:1232–1240. 10.1104/pp.107.106377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Malloy PW, Paul CL. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A. 89:1827–1831. 10.1073/pnas.89.5.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R. 2008. Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9:371. 10.1186/1471-2105-9-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagen C, Rojas MR, Kon T, Gilbertson RL. 2008. Recovery from Cucurbit leaf crumple virus (family Geminiviridae, genus Begomovirus) infection is an adaptive antiviral response associated with changes in small viral RNAs. Phytopathology 98:1029–1037. 10.1094/PHYTO-98-9-1029 [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Negrete EA, Carrillo-Trip J, Rivera-Bustamante RF. 2009. RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 83:1332–1340. 10.1128/JVI.01474-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brough CL, Gardiner WE, Inamdar N, Zhang XY, Ehrlich M, Bisaro DM. 1992. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 18:703–712. 10.1007/BF00020012 [DOI] [PubMed] [Google Scholar]
- 53.Ermak G, Paszkowski U, Wohlmuth M, Scheid OM, Paszkowski J. 1993. Cytosine methylation inhibits replication of African cassava mosaic virus by two distinct mechanisms. Nucleic Acids Res. 21:3445–3450. 10.1093/nar/21.15.3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15:1494–1500. 10.1016/j.cub.2005.07.024 [DOI] [PubMed] [Google Scholar]
- 55.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38:721–725. 10.1038/ng1804 [DOI] [PubMed] [Google Scholar]
- 56.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park H-S, Vazquez F, Robertson D, Meins F, Hohn T, Pooggin MM. 2006. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34:6233–6246. 10.1093/nar/gkl886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25:3347–3356. 10.1038/sj.emboj.7601217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deleris A, Gallego-Bartolome J, Bao J, Kasschau K, Carrington JC, Voinnet O. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68–71. 10.1126/science.1128214 [DOI] [PubMed] [Google Scholar]
- 59.Cañizares MC, Lozano-Durán R, Canto T, Bejarano ER, Bisaro DM, Navas-Castillo J, Moriones E. 2013. Effects of crinivirus coat protein-interacting plant protein SAHH on post-transcriptional RNA silencing and its suppression. Mol. Plant Microbe Interact. 26:1004–1015. 10.1094/MPMI-02-13-0037-R [DOI] [PubMed] [Google Scholar]
- 60.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW-L, Lagrange T, Pikaard CS, Jacobsen SE. 2006. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126:93–106. 10.1016/j.cell.2006.05.032 [DOI] [PubMed] [Google Scholar]
- 61.Li CF, Henderson IR, Song L, Federoff N, Lagrange T, Jacobsen SE. 2008. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet. 4:e27. 10.1371/journal.pgen.0040027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akbergenov R, Si-Ammour A, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins F, Jr, Hohn T, Pooggin MM. 2006. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 34:462–471. 10.1093/nar/gkj447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator V3: a reference expression database for meta-analysis of transcriptomes. Adv. Bioinformatics 2008:420747. 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pelligrini M, Jacobsen SE. 2008. Shotgun bisuphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219. 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523–536. 10.1016/j.cell.2008.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zemach A, Kim MY, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer S, Zilberman D. 2013. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153:193–205. 10.1016/j.cell.2013.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK. 2013. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162:116–131. 10.1104/pp.113.216481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeske H, Lutgemeier M, Preiss W. 2001. DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J. 20:6158–6167. 10.1093/emboj/20.21.6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Preiss W, Jeske H. 2003. Multitasking in replication is common among geminiviruses. J. Virol. 77:2972–2980. 10.1128/JVI.77.5.2972-2980.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paprotka T, Deuschle K, Metzler V, Jeske H. 2011. Conformation-selective methylation of geminivirus DNA. J. Virol. 85:12001–12012. 10.1128/JVI.05567-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. 2004. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 14:1214–1220. 10.1016/j.cub.2004.06.055 [DOI] [PubMed] [Google Scholar]