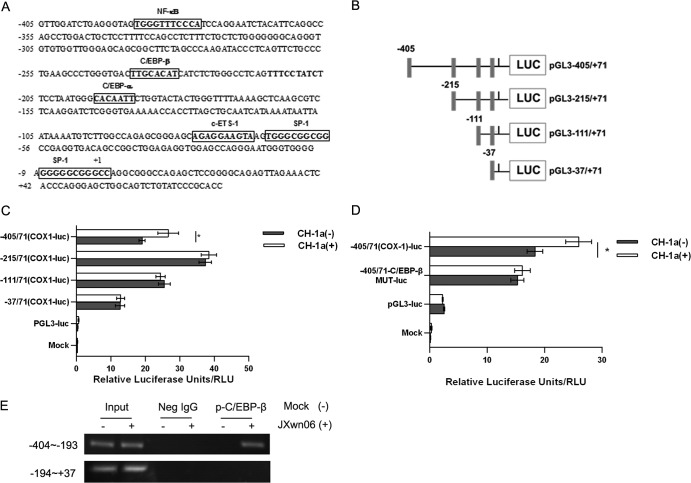
FIG 5.
Cloning and sequencing of the porcine COX-1 promoter and mapping of an essential PRRSV-responsive region. (A) Cloning and sequence analysis of the 476-bp porcine COX-1 promoter. The putative transcription factor binding sites are shown in boxes; the positions of the putative regulatory motifs are relative to the transcription initiation site. (B) Schematic representation of the truncated COX-1 promoter in a basic luciferase (LUC) expression vector (pGL3) (numbers indicate base positions). Black rectangles, translation initiation. (C) Marc-145 cells were transfected with 2 μg of each constructed plasmid for 24 h, and then cells were infected with or without the CH-1a strain of virus (MOI, 2). At 24 h postinfection, cells were harvested for luciferase activity analysis. (D) Effect of mutation in the putative C/EBP-β region on the COX-1 promoter activity induced by PRRSV. Marc-145 cells were transfected with 2 μg of the constructed mutant promoter for 24 h, and then cells were infected with or without CH-1a virus (MOI of 2). At 24 h postinfection, cells were harvested to determine luciferase activity. (E) p-C/EBP-β and COX-1 promoter interaction. PAMs were incubated with HP-PRRSV strain JXnw06 (MOI, 2) for 6 h, and a ChIP assay was then performed as described in Materials and Methods. Data are means ± SEMs from three independent experiments. Differences were evaluated by Student's t test. (*, P < 0.05).