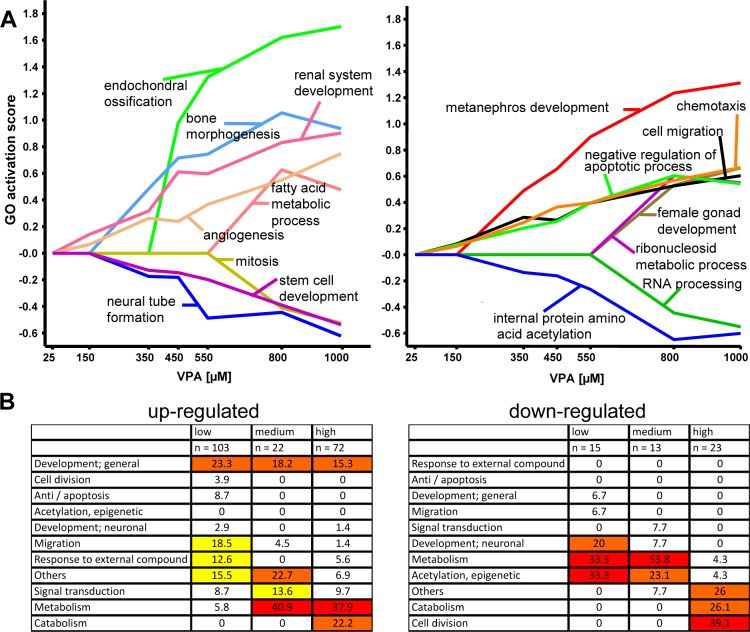
Figure 6.
Concentration-dependent regulation of GOs and of superordinate cell biological processes. The test system was exposed to various concentrations of VPA for 6 days. Then, transcriptome analysis was performed, and differentially expressed genes were identified as in Figure 1. The data sets were examined for overrepresentation of any GO, and GO-based concentration–response profiles were generated as detailed in Table S8 (Supporting Information). Examples for transcriptome activation responses on the level of GO are shown. (A) Quantitative GO activation score was calculated by multiplying the percentage of genes within the GO that was found to be significantly regulated with the average fold change of these regulations. Positive values reflect oGO among up-regulated genes, and negative values indicate oGO among down-regulated genes. The names of the example GOs are indicated in the figures. Note the different concentrations at which a GO is first turned on and the different types of concentration–response behavior at high concentrations. (B). All GO profiles among up- and down-regulated genes were sorted, and the following three groups were selected for further analysis: low (GO found to be up-regulated at all VPA concentrations starting from 150 μM, clusters 0++++++ and 00+++++); intermediate (GO found to be up-regulated at all VPA concentrations higher than 350 μM, clusters 000++++ and 0000+++); and high (GO found to be up-regulated only at 800 and 1000 μM, clusters 00000++ and 000000+). Then, the GO were assigned to superordinate cell biological processes (e.g., migration and adhesion), and we calculated how many of all oGO in the low–medium–high groups belonged to the biological processes. For easier overview, a color code was applied with no color for values <10%, yellow for 10–20%, orange for 20–30%, and red for >30%.