Figure 4.
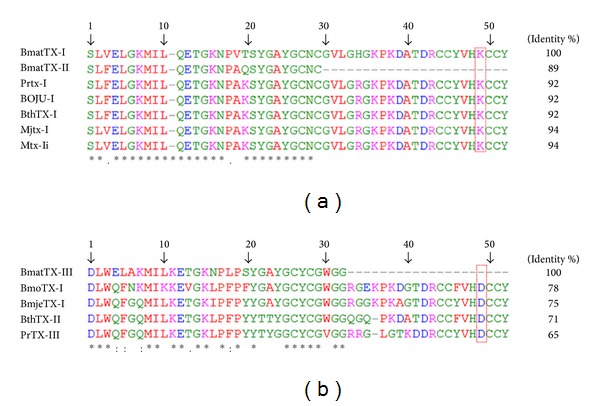
Comparison of the N-terminal sequence of the snake's PLA2s with the PLA2s isolated from B. mattogrossensis venom. N-terminal sequencing of the PLA2s obtained by Edman [13] degradation and alignment done with CLUSTALW2 expressed as % of similarity. (a) BmatTX-I, BmatTX-II with Lys49 residues in marked sequences. (b) BmatTX-III with Asp49 residues in marked sequences. Sequences used for the alignment and their respective access numbers: BthTX-I (gi:51890398); MjTX-I (gi:17368325); Mtx-II (gi:390981003); PrTX-I (gi:190016174); BOJU-I (gi:209572966); BthTX-II (gi:1171971); PrTX-III (gi:90016174); BmjeTX-I (gi:313471399); BmoTX-I (gi:221272396). BthTX-I (Lys49), BthTX-II (Asp49), and BOJU-I (Lys49): isolated from the venom of B. jararacussu; MjTX-I (Lys49), BmoTX-I (Asp49), and BmjeTX-I (Asp49): isolated from the venom of B. moojeni; Mtx-II (Lys49): isolated from the venom of B. brazili; PrTX-I (Lys49) and PrTX-III (Asp49): isolated from the venom of B. pirajai; the “∗” was used to indicate the amino acid residues that are the same between the sequences.
