FIG 3 .
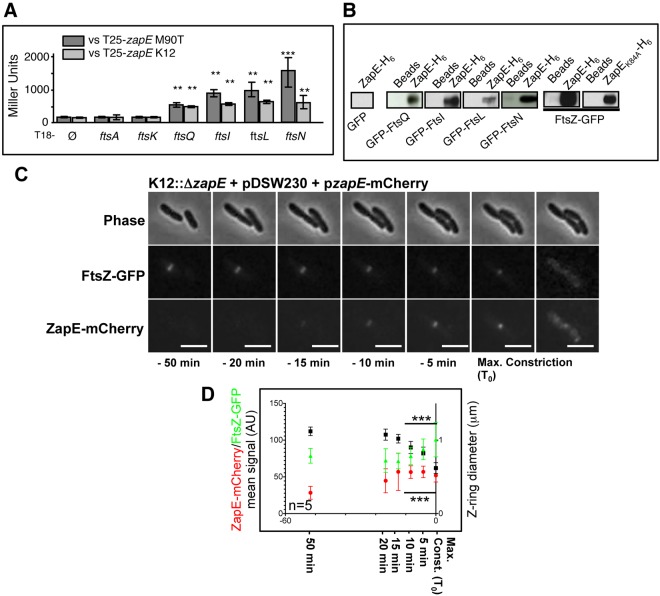
ZapE is a cytoplasmic protein which interacts with FtsZ in vitro and in vivo. ZapE recruitment correlates with Z-ring constriction. (A) BACTH analysis was performed using the T25-zapE K-12 (E. coli K-12) or T25-zapE M90T (Shigella M90T) versus T18-plasmid constructs with indicated genes. Results are expressed in Miller units and were averaged from three independent experiments. Error bars show the standard deviations (SD). Comparing average activity to that of the T18 negative control, ** indicates P < 0.01 and *** indicates P < 0.001 (Student’s t test). (B) Pulldown assay with K-12 ZapE-H6 and GFP or GFP-tagged proteins in E. coli lysates. The interaction between E. coli FtsZ-GFP (pDSW230) and E. coli ZapE-H6 and ZapEK84A-H6, respectively, was analyzed using a His pulldown assay. (C) Expression and localization of FtsZ-GFP and ZapE-mCherry during a cell division. Time-lapse observation was performed on an LB-agar pad at 30°C, using a 200 M Axiovert epifluorescence microscope (Zeiss). Image acquisition was performed every 3 min (see also Movie S1 and Fig. S4C in the supplemental material for raw fluorescence quantification). This result is representative of five individual observations from three independent experiments. Bars are 2 µm. Max, maximum. (D) Relationship between, respectively, ZapE-mCherry and the FtsZ-GFP mean signal (AU) (K12::ΔzapE pzapE-mCherry) and the Z-ring diameter (pDSW230). Mean FtsZ-GFP and ZapE-mCherry fluorescent signals are represented in Fig. S4C in the supplemental material. n = 5 independent observations; error bars show the SD. *** indicates P < 0.001 (Student’s t test). Max. Const., maximum constriction.