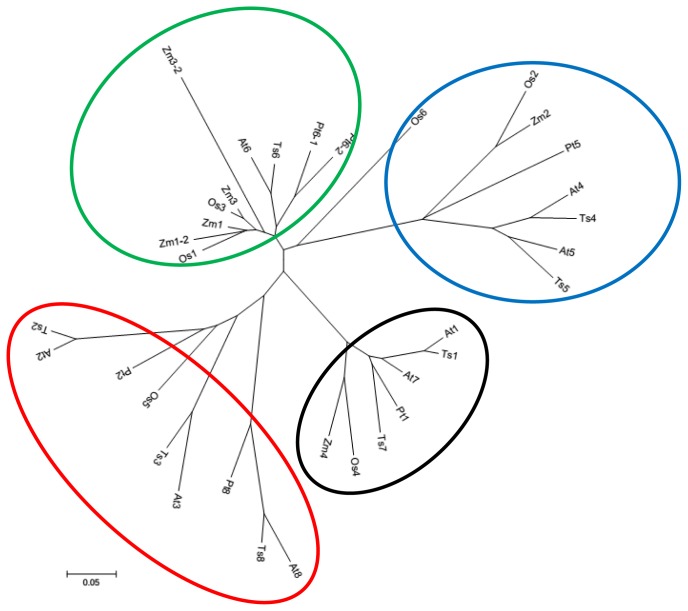
Figure 3.
Phylogenetic analyses of thirty four plant glutathione peroxidase (GPX) proteins. The tree was constructed using the neighbor-joining method of CLUSTALW, with 1000 bootstraps, and the bar indicates 0.05 substitutions per site. Each ellipse shows a clade. Abbreviations of plant species: Ts, Thellungiella salsuginea; At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays, Pt, Populus trichocarpa. The plant GPXs include At1 (At2g25080), At2 (At2g31570), At3 (At2g43350), At4 (At2g48150), At5 (At3g63080), At6 (At4g11600), At7 (At4g31870), At8 (At1g63460), Pt1 (POPTR_0006s28120), Pt2 (POPTR_0007s02160), Pt5 (POPTR_0014s13490), Pt6-1 (POPTR_0001s09270), Pt6-2 (POPTR_0003s12620), Pt8 (POPTR_0001s09280), Os1 (Os04g0556300), Os2 (Os03g0358100), Os3 (Os02g0664000), Os4 (Os06g0185900), Os5 (Os11g18170), Os6 (A3AYS5_ORYSJ), Zm1 (Q6JAH6_MAIZE), Zm1-2 (B6SU31_MAIZE), Zm2 (B6T5N2_MAIZE), Zm3 (B4FRF0_MAIZE), Zm3-2 (AC204541), and Zm4 (B6U7S4_MAIZE).