Fig. 3.
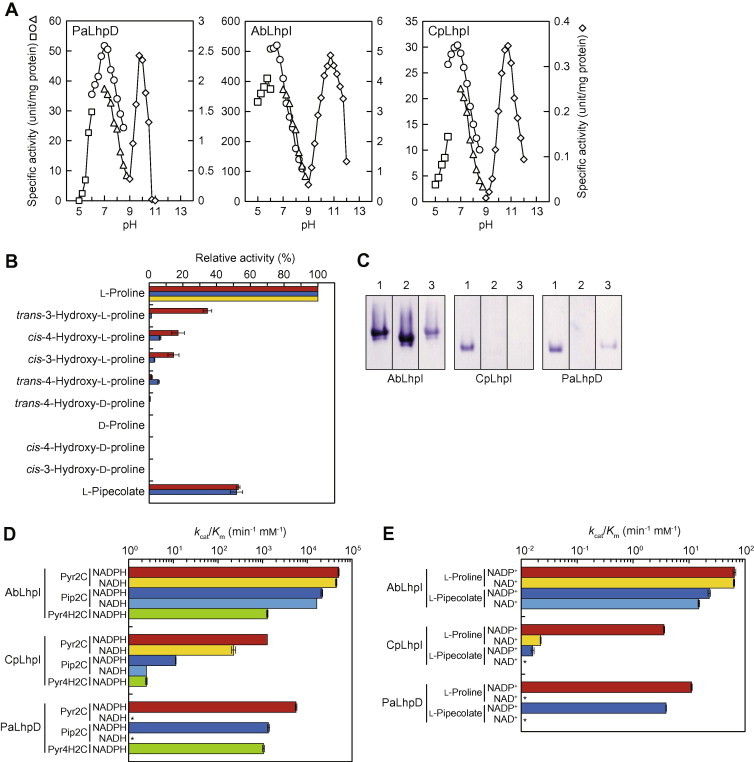
Enzymatic properties of Pyr2C reductase. (A) Effect of pH on the activity. Left panel, PaLhpD; middle panel, AbLhpI; right panel, CpLhpI. 50 mM acetate-NaOH (pH 4.0–6.0) (square), 50 mM potassium phosphate (pH 6.0–8.5) (circle), 50 mM Tris–HCl (pH 7.0–9.0) (triangle) for the reduction of Pyr2C (left axis), and 50 mM glycine–NaOH (diamond) for the oxidization of l-proline (right axis), instead of 50 mM Tris-HCl (pH 8.0) under standard assay conditions. (B) Substrate specificity of NADP+-dependent oxidization reaction. PaLhpD, red bar; AbLhpI, blue bar; CpLhpI, yellow bar. The assay was performed with standard assay solution containing the indicated substrate (10 mM). Relative values were expressed as percents of the values obtained in l-proline (means ± SD, n = 3). (C) Zymogram staining. Five micrograms of each of purified protein (left panel, AbLhpI; middle panel, CpLhpI; right panel, PaLhpD) was applied on 10% (w/v) gel. After electrophoresis, the gel was soaked in staining solution in the presence of substrates and coenzymes as follows (each of 10 mM): lane 1, l-proline and NADP+; lane 2, l-proline and NAD+; lane 3, l-pipecolate and NADP+. (D) Catalytic efficiency (kcat/Km) of NADPH-dependent reduction (left panel) and NADP+-dependent oxidization reactions (right panel). Asterisks indicate no (or trace) activity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
