Fig. 5.
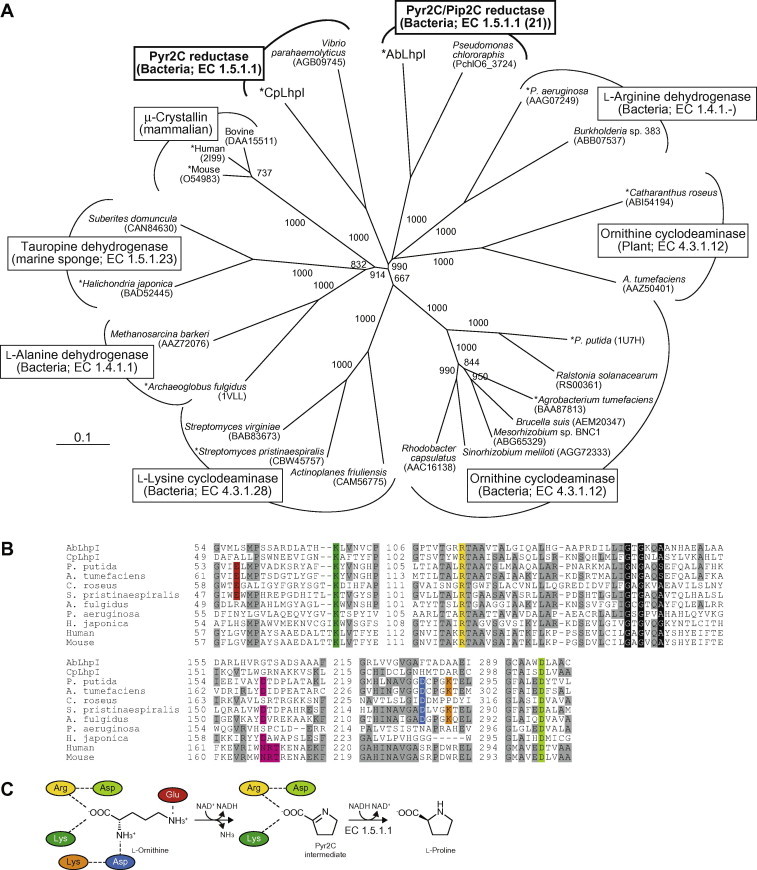
(A) Phylogenetic tree of OCD/μ-crystallin protein family. The number on each branch indicates the bootstrap value. Proteins with asterisks were used for B. (B) Partial multiple sequence alignment of deduced amino acid sequences of Pyr2C reductases. Binding sites for a carboxyl group of substrate, lysine, arginine and aspartate, are shaded in green, yellow and light-green, respectively. Aspartate and glutamate residues, related to the cyclization by OCD and LCD, are shaded in red and blue, respectively. Amino acid residues interacting with 2′- and 3′-functional groups in the ribosyl moiety of NAD+(H) and NADP+(H) are shaded in pink. Coenzyme-binding motif of Rossmann-fold are shown as white letters in black boxes. Gray-shaded letters indicate highly conserved amino acid residues. (C) Schematic diagram showing the interactions of l-ornithine, Pyr2C, and nearby residues in OCD. Color of residues correspond to B. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
