Figure-1.
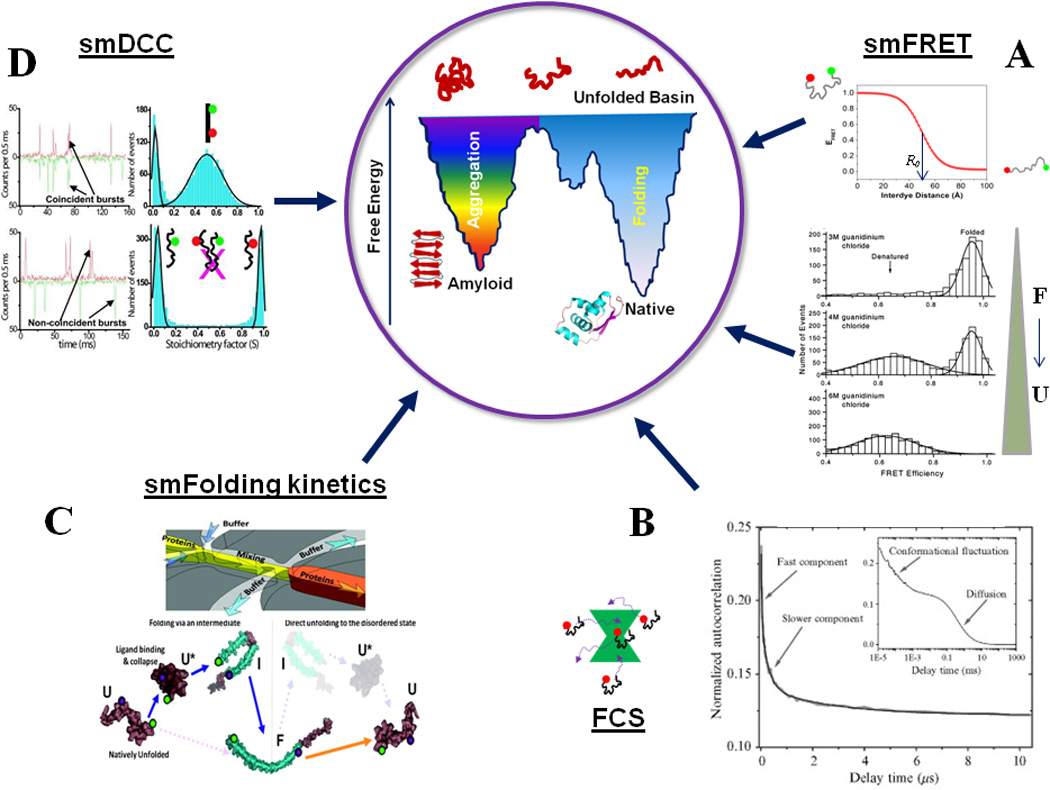
An overview of single-molecule fluorescence methods for protein folding studies. (A) smFRET can detect individual subpopulations of protein conformations in a heterogeneous structural ensemble by means of FRET efficiency, which is a function of inter-dye distance (Eqn. 1). smFRET histograms showing denaturant induced unfolding of a two-state folder, CI2. Adapted from Deniz et al., Proc. Natl. Acad. Sci. USA (2000), 97:5179 50. (B) FCS can be employed to detect slow and fast conformational dynamics of proteins: shown here are the autocorrelation curves for the NM domain of Sup35. Adapted from Mukhopadhyay et al. Proc. Natl. Acad. Sci., USA (2007) 104:2649 29. (C) Shows a microfluidic mixer which can be coupled with smFRET detection to study single-molecule folding and unfolding pathways of proteins. The bottom shows a schematic of the observed pathways for α-synuclein. Adapted from Gambin et al., Nature Methods (2011) 8:239 102. (D) Shows the application of smDCC for directly detecting lack of aggregation of the NM domain of Sup35 under experimental conditions. Adapted from Mukhopadhyay et al., Proc. Natl. Acad. Sci. USA (2007) 104:2649 29. Aggregation would be signaled by coincident bursts, which could be analyzed for further quantification. The concepts behind the central cartoon showing a double-funnel for protein folding and aggregation are discussed in 146. The native state protein structure at the bottom of the folding funnel is a cartoon for MJ0366 (pdb ID: 2EFV).
