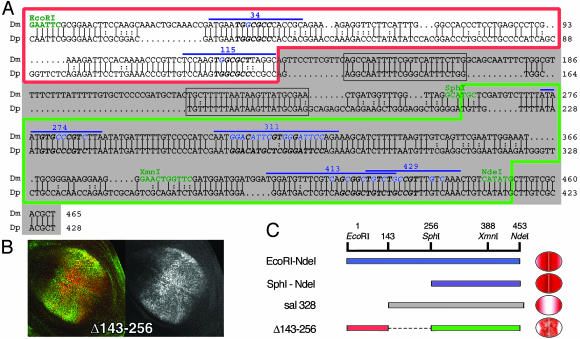
Fig. 5.
salE/Pv enhancer is conserved in D. pseudoobscura. (A) Alignment of D. melanogaster (Dm) and D. pseudoobscura (Dp) salE/Pv enhancers. Only the EcoRI–NdeI region of the enhancer is shown. Relevant restriction enzymes are shown in green. Oligonucleotides used in this analysis are indicated with a blue line. The nucleotides mutated in the different experiments are shown in blue. The red line boxes the repressor region, and the green line boxes the region important for activation. sal328 fragment is shaded in gray. The conserved regions eliminated in the mutant salE/Pv-Δ143–256 are boxed in gray. Italics and bold characters indicate consensus sequences for Brk, Med, or Sd. (B) Expression of Sal (green) and β-gal (red) in third-instar wing imaginal discs driven by the salE/Pv-Δ143–256 fragment. The corresponding red channel is in black and white. (C) Schematic representation of the EcoRI–NdeI fragment, which drives the reporter expression in the central part of the wing blade. When the EcoRI–SphI fragment is removed, the expression expands laterally (SphI–NdeI). The normal domain of expression is recovered when the repressor sites are added, albeit the levels of expression are heterogeneous (Δ143–256). However, the elimination of the repressor sites produces a change in the specificity of the enhancer that now drives reporter expression only in the lateral regions of the wing blade (sal328).