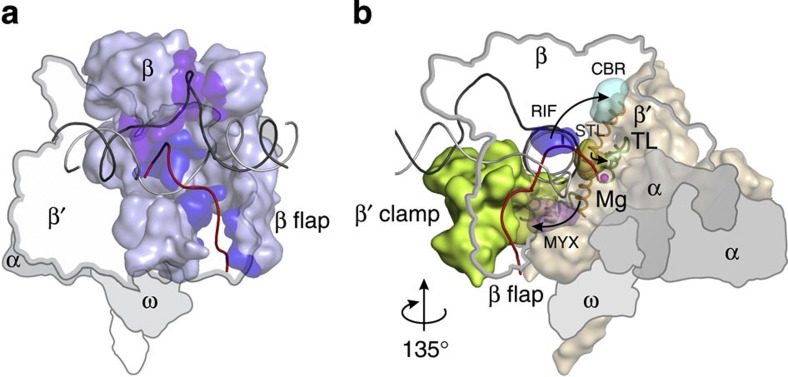
Figure 6. Antibiotic-binding sites outline an allosteric path mediating regulation of transcription elongation.
(a) The β subunit (light blue) interacts with nucleic-acid determinants implicated in regulation of transcription elongation53,54,55,56: the nascent RNA57 (red, blue β surface) and unpaired non-template DNA52,58 (black, purple β surface). β′ subunit is depicted as a contour, and α and ω subunits as grey outlines. (b) Allosteric effects (black arrows) travel through rifampicin (RIF, sterically59 and allosterically60 inhibits transcription initiation) -binding site (blue surface, amino-acid substitutions alter RNAP pausing propensities26) in the β subunit. CBR (inhibits transcription elongation by promoting pausing)-binding site (cyan surface, amino-acid substitutions lead to pausing defects) outlines the interface where allosteric effects are transferred to the BH (orange cartoon). STL (inhibits transcription elongation by restricting TL and BH conformations28,32) -binding site (yellow surface) marks the region where conformational changes in BH modulate the stability of the folded TL (green cartoon). Myxopyronin (restricts mobility of the β′ clamp61,62,63 and alters position of template DNA strand64) binds to the flexible region (pink surface, switch 1 as green cartoon on the background) that controls movement of β′ clamp (green surface) and conformation of the template DNA strand (grey cartoon). β subunit is depicted as a contour, and α and ω subunits as grey outlines. The figure was prepared using PyMOL Molecular Graphics System, Version 1.6.0.0; Schrödinger, LLC. The sources of atomic coordinates are listed in Supplementary Table 3.