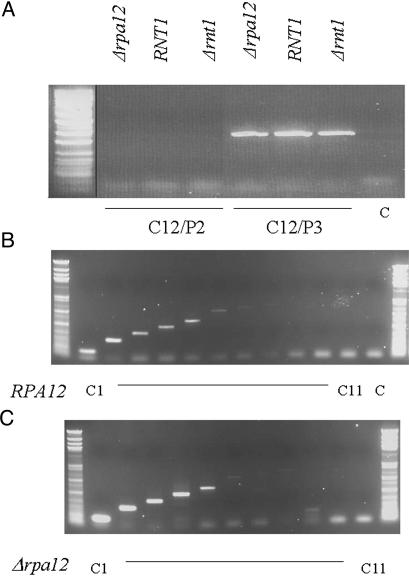
Fig. 3.
RT-PCR analysis of steady-state rRNA generated in wild-type, Δrnt1, and Δrpa12 strains. (A) RT-PCR across the Pol I transcription start site reveals no detectable steady-state transcription reading into the Pol I promoter region in any strain tested. RT and PCR primers used are indicated, as are their positions relative to the rDNA repeat (Fig. 1 A). PCR products are resolved by agarose gel electrophoresis. (B and C) RT-PCR analysis beyond the normal Reb1p terminator is shown for isogenic wild-type and Δrpa12 strains. Faint signals are detected up to cDNA primer C5, which maps to rDNA nucleotides 7598–7618, but the same profile is found with both strains. cDNAs were generated with primers C1–C11 and amplified by using primer P1.