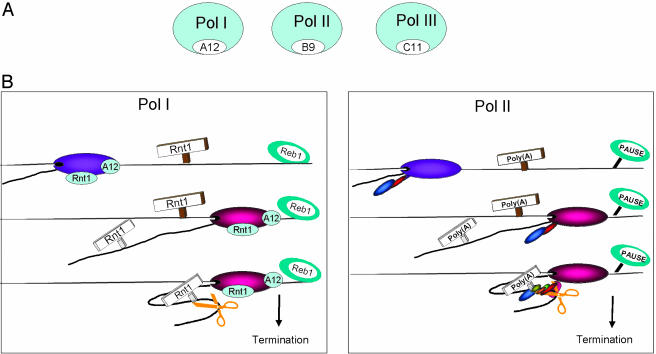
Fig. 4.
Homologous mechanisms of transcriptional termination for Pol I and II. (A) Homologous small subunits (white shading) in all three polymerase classes. (B) Model showing marked similarity in the mechanisms of Pol I and II termination. Polymerases before the terminal cleavage site are colored purple and after, red, to indicate possible conformational change. The Pol II CTD tail with associated 3′ end processing factors is indicated. Both Pol I and II nascent transcripts are predicted to fold around and interact with paused polymerase. The associated 3′ processing factors will then mediate 3′ end cleavage (orange scissors). This process is facilitated by polymerase pausing, which in turn will induce transcriptional termination. For Pol I, this depends on Reb1p and may require interactions with the Pol I subunit, Rpa12p.