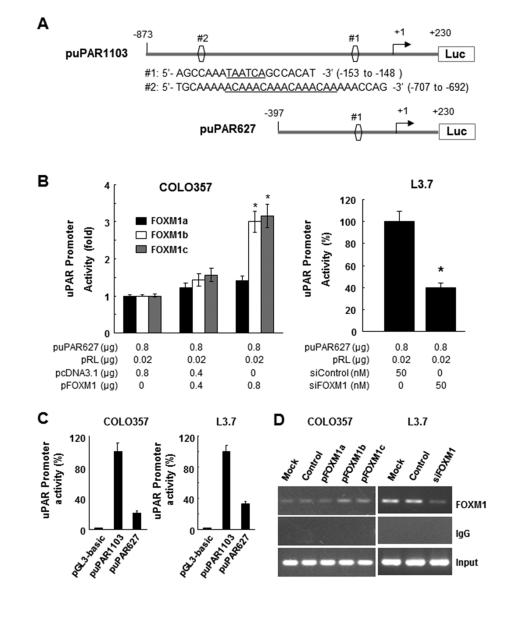
Figure 5.
Direct binding of FOXM1 to the uPAR promoter. A, sequences and positions of putative FOXM1-binding elements on the uPAR promoter puPAR1103 (sites #1 and #2) and a deletion mutation of the uPAR promoter puPAR627 (site #2). B, COLO357 cells were co-transfected with 0.8 mg of the uPAR promoter luciferase construct puPAR1103 and 0-0.8 mg of pcDNA3.1-FOXM1a (pFOXM1a), pcDNA3.1-FOXM1b (pFOXM1b), pcDNA3.1-FOXM1c (pFOXM1c), or pcDNA3.1 (left), whereas L3.7 cells were co-transfected with 0.8 μg of puPAR1103 and 50 nmol/L siFOXM1 or control siRNA (siControl; right). The promoter activity in the cells was measured using a dual luciferase assay kit. *P < 0.01. C, COLO357 and L3.7 cells were co-transfected with pGL3-basic, puPAR1103, or puPAR627. The promoter activity in the cells was assessed using a dual luciferase assay kit. D, ChIP assay. Chromatins were isolated from COLO357 cells transfected with pcDNA3.1-FOXM1a, pcDNA3.1-FOXM1b, pcDNA3.1-FOXM1c, or pcDNA3.1, and binding of FOXM1 to the uPAR promoter was analyzed using a specific anti-FOXM1 antibody and oligonucleotides flanking the uPAR promoter regions containing putative FOXM1-binding sites as described in Materials and Methods (left). Similar ChIP assays were conducted using chromatins isolated from L3.7 and L3.7-siFOXM1 cells (right). Normal IgG was used as a control, and 1% of the total cell lysates was subjected to PCR analysis before immunoprecipitation (input control).