Table 2. Top 10 PHX genes from genomes of 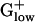 bacteria.
bacteria.
|
E(g)*
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | BACSU | BACHA | LISIN | LISMO | LACLA | STRPY | STRPN | STAAU | CLOAC | CLOPE |
| RPs | ||||||||||
| L1 (rplA) | 1.91 | 1.55 | 2.05 | 2.03 | 2.00 | 1.95 | 2.11 | 2.12 | 1.18 | 1.71 |
| L2 (rplB) | 2.02 | 1.69 | 1.92 | 1.85 | 2.09 | 2.16 | 2.33 | 1.70 | 1.40 | 1.52 |
| L3 (rplC) | 1.78 | 1.60 | 1.44 | 1.42 | 1.71 | 1.73 | 1.87 | 1.59 | 1.30 | 1.85 |
| L4 (rplD) | 1.98 | 1.67 | 1.69 | 1.73 | 1.70 | 1.71 | 1.91 | 1.46 | 1.23 | 1.24 |
| L5 (rplE) | 1.92 | 1.35 | 1.34 | 1.36 | 1.79 | 1.69 | 2.17 | 1.79 | 1.52 | 1.72 |
| L14 (rplN) | 1.98 | 1.43 | 1.30 | 1.37 | 1.51 | 1.69 | 1.89 | 1.70 | 1.08 | 1.24 |
| L17 (rplQ) | 1.87 | 1.51 | 1.69 | 1.72 | 2.31 | — | 1.83 | 2.03 | 1.18 | 1.23 |
| L19 (rplS) | 1.84 | 1.62 | 1.54 | 1.60 | 1.90 | 1.76 | 1.99 | 1.71 | 1.38 | 1.55 |
| L20 (rplT) | 1.57 | 1.54 | 1.38 | 1.42 | 1.57 | 1.72 | 2.15 | 1.80 | 1.24 | 1.96 |
| S1 (rpsA) | 1.20 | (1.01) | (0.90) | 1.17 | 2.14 | 2.32 | 2.31 | (0.80) | (0.74) | (0.76) |
| S2 (rpsB) | 1.84 | 1.47 | 1.72 | 1.67 | 2.37 | 2.03 | 2.30 | 2.09 | 1.33 | 1.58 |
| S3 (rpsC) | 1.87 | 1.48 | 1.69 | 1.70 | 2.17 | 1.95 | 2.10 | 1.83 | 1.21 | 1.99 |
| S4 (rpsD) | 1.94 | 1.61 | 1.84 | 1.82 | 2.09 | 2.12 | 2.33 | 2.11 | 1.19 | 1.67 |
| S9 (rpsI) | 1.51 | 1.53 | 1.86 | 1.87 | 1.52 | 1.92 | 1.87 | 2.36 | 1.42 | 1.46 |
| S13 (rpsM) | 2.02 | 1.38 | 1.93 | 1.86 | 1.59 | 1.72 | 1.95 | 1.86 | 1.27 | 1.23 |
| TFs | ||||||||||
| Translation elongation factor G (fus) | 2.34 | 1.77 | 2.34 | 2.31 | 2.46 | 2.16 | 2.47 | 2.64 | 1.45 | 2.08 |
| Translation elongation factor Ts (tsf) | 1.75 | 1.44 | 1.98 | 2.33 | 1.41 | 1.87 | 2.35 | 2.11 | 1.31 | 1.62 |
| Translation elongation factor Tu (tuf) | 1.97 | 1.50 | 2.00 | 1.92 | 1.91 | 1.96 | 2.06 | 2.09 | 1.14 | 1.42 |
| 1.42 | ||||||||||
| Translation initiation factor IF-2 (infB) | (0.79) | (0.77) | 1.06 | 1.23 | 1.65 | (0.56) | 1.04 | 1.00 | (1.04) | 2.04 |
| RNA polymerase β-subunit (rpoB) | 1.49 | 1.19 | 2.37 | 2.19 | 1.87 | 1.72 | 1.83 | 1.67 | (0.98) | 1.62 |
| RNA polymerase β′-subunit (rpoC) | 1.76 | 1.54 | 2.39 | 2.26 | 2.02 | 1.72 | 1.77 | 1.55 | 1.19 | 1.53 |
| GTP-binding protein TypA/BipA | 1.11 | (0.78) | 1.91 | 2.09 | 1.95 | 1.45 | 1.98 | (0.97) | 1.18 | 1.45 |
| Chaperones | ||||||||||
| HSP 60 (groEL) | 1.87 | 1.79 | 1.91 | 1.89 | 1.23 | (0.86) | 1.08 | 1.19 | 1.45 | (0.71) |
| HSP 70 (dnaK) | 1.83 | 1.14 | 2.25 | 2.17 | 2.08 | 2.25 | 2.43 | 2.21 | 1.38 | 2.19 |
| Trigger factor (tig) | 1.85 | 1.53 | 2.02 | 1.81 | 1.86 | 1.55 | 2.55 | 2.02 | 1.42 | 1.60 |
| Glycolysis | ||||||||||
| Glucose-6-phosphate isomerase (pgi) | (0.92) | (0.93) | 1.67 | 1.77 | 2.13 | 1.57 | 2.46 | 1.33 | 1.28 | 1.74 |
| Fructose-1,6-bisphosphate aldolase (fba) | 1.99 | 1.20 | 1.71 | 1.70 | 2.08 | 2.07 | 2.15 | 2.00 | 1.20 | 1.52 |
| (0.59) | (0.83) | (0.67) | (0.59) | (0.86) | (0.71) | |||||
| (etc.) | (etc.) | |||||||||
| Glyceraldehyde-3-phosphate dehydrogenase | 1.80 | 1.53 | 1.77 | 1.72 | 1.95 | 2.13 | 2.08 | 2.12 | 1.27 | 1.48 |
| (gap) | (0.48) | (0.70) | 1.04 | (0.69) | ||||||
| Phosphoglycerate kinase (pgk) | 1.34 | 1.09 | 2.08 | 2.02 | 2.30 | 2.18 | 2.24 | 1.86 | 1.34 | 1.65 |
| Phosphoglycerate mutase (gpmA) | (0.70) | — | (0.93) | (1.04) | 2.27 | 1.88 | 1.94 | (0.79) | (0.95) | — |
| Phosphoglycerate mutase (2.3-bisphosphoglycerate-independent) (pgm) | 1.08 | (0.88) | 1.97 | 1.98 | — | — | — | 1.15 | 1.29 | 1.80 |
| Enolase (eno) | 1.92 | 1.61 | 1.83 | 1.92 | 1.96 | 2.17 | 2.24 | 2.12 | 1.26 | 1.51 |
| (0.49) | ||||||||||
| Pyruvate kinase (pykA) | 1.18 | (0.76) | 2.38 | 2.17 | 2.25 | 2.04 | 2.28 | 1.70 | 1.29 | 1.68 |
| (0.83) | ||||||||||
| 1.29 | ||||||||||
| PTS | ||||||||||
| Phosphotransferase enzyme IIC component | (0.57) | (0.57) | 2.06 | 2.05 | 1.32 | (0.55) | (1.01)† | — | 1.24 | — |
| (cellobiose-specific) | (0.46) | 1.16 | 1.32 | 1.10 | (0.60) | |||||
| (0.45) | (etc.) | (etc.) | (0.47) | (0.49) | ||||||
| PTS component IID (mannose-specific) (manN) | — | — | 1.26 | 1.47 | 1.85 | 2.15 | 1.54 | — | — | 1.27 |
| 1.23 | 1.09 | 1.04 | ||||||||
| PTS system, IIABC components‡ | — | — | — | — | — | (0.60) | 2.43 | — | — | (0.80) |
| Pyruvate dehydrogenase, pyruvate oxidase | ||||||||||
| Dihydrolipoamide acelyltransferase component of pyruvate dehydrogenase complex (pdhC) | 2.05 | 1.48 | 1.79 | 1.81 | (0.94) | — | — | 1.43 | — | — |
| Dihydrolipoamide dehydrogenase E3-subunit of pyruvate dehydrogenase (pdhD) | 2.14 | 1.55 | 1.98 | 1.80 | 1.09 | — | — | 2.05 | — | — |
| Pyruvate oxidase | (0.47) | — | (0.86) | 1.03 | (0.47) | — | 2.48 | (0.66) | — | — |
| Fermentation and anaerobic respiration | ||||||||||
| Pyruvate formate-lyase (formate acetyltransferase) (pflB, pflA) | — | — | 2.40 | 2.49 | 1.78 | 2.32 | 2.87 | 2.09 | (0.98) | 1.92 |
| 1.98 | 2.09 | |||||||||
| l-lactate dehydrogenase (ldh) | (0.76) | (0.69) | 1.63 | 1.53 | 2.26 | 1.72 | 2.30 | (0.90) | (0.94) | (0.78) |
| (0.45) | (0.88) | (0.80) | ||||||||
| (0.44) | ||||||||||
| Alcohol-acetaldehyde dehydrogenase (adhE) | — | — | 2.08 | 2.14 | 1.44 | — | 2.22 | (0.56) | — | 2.00 |
| β-hydroxybutyryl-CoA dehydrogenase, NAD-dependent (CAC2708) | (0.52) | (0.81) | — | — | — | — | — | — | 1.44 | 1.55 |
| (0.71) | ||||||||||
| (0.68) | ||||||||||
| Butyryl-CoA dehydrogenase (CAC2711) | (0.56) | (0.66) | — | — | — | — | — | — | 1.44 | 1.70 |
| (0.48) | (0.63) | (0.66) | ||||||||
| (0.41) | (0.62) | |||||||||
| Pyruvate ferredoxin oxidoreductase (CAC2229, CPE2061) | — | — | (0.51) | (0.45) | (0.40) | — | — | — | 1.43 | 2.05 |
| 1.07 | ||||||||||
| Amino acid blosynthesis | ||||||||||
| Ketol-acid reductoisomerase (ilvC) | 1.03 | 1.59 | 1.16 | 1.27 | (0.79) | — | 2.11 | (1.06) | 1.04 | — |
| Methionine synthase (metE) | (0.72) | (0.88) | (0.56) | (0.61)† | (0.44) | — | 2.44 | 1.34 | — | — |
| Transporters | ||||||||||
| Permease of the Na+:galactoside symporter family (CAC0694, yjmB) | (0.46) | — | — | — | — | — | — | — | 1.47 | — |
| ABC transporter, substrate-binding protein (SP0092) | — | (0.63) | (0.83) | (0.82) | (0.68) | (0.80) | 2.73 | — | — | 1.54 |
| Oligonucleotide ABC transporter (BH0031) | — | 1.78 | — | — | — | — | — | — | — | — |
| (0.66) | ||||||||||
| (etc.) | ||||||||||
| Oligopeptide ABC transporter (CAC3643) | (0.58) | (0.76) | (0.69) | (0.75) | (0.69) | — | — | (0.51) | 1.42 | (0.65) |
| (0.49) | (1.06) | |||||||||
| Glycogen degradation | ||||||||||
| Glycogen phosphorylase (glgP) | (0.39) | (0.52) | — | — | (0.42) | 1.27 | 2.88 | — | (0.90) | (0.57) |
| (0.43) | ||||||||||
| Phosphoglucomutase | (0.54) | (0.64) | (0.44) | (0.56) | — | (0.84) | 2.46 | — | — | — |
| Other | ||||||||||
| P60 extracellular protein, invasion-associated (iap) | — | — | 2.16 | 1.76 | — | — | — | — | — | — |
| Hypothetical protein (CPE1232) | — | — | — | — | — | — | — | — | 1.35 | 2.00 |
| Hypothetical protein (CPE1233) | — | — | — | — | — | — | — | — | 1.30 | 1.90 |
Included are all genes ranking among the top 10 PHX genes in any of the eight genomes (underlined) and their homologs in other genomes even if they are not among the top 10.
Numbers in parentheses indicate the gene is not PHX; —, the gene does not have a homolog in the genome; etc, more than three homologs
PA gene
All genomes have homologs of PTS system IIABC components but of lower similarity (≈30%). Those listed exhibit high mutual similarity (>50%)
