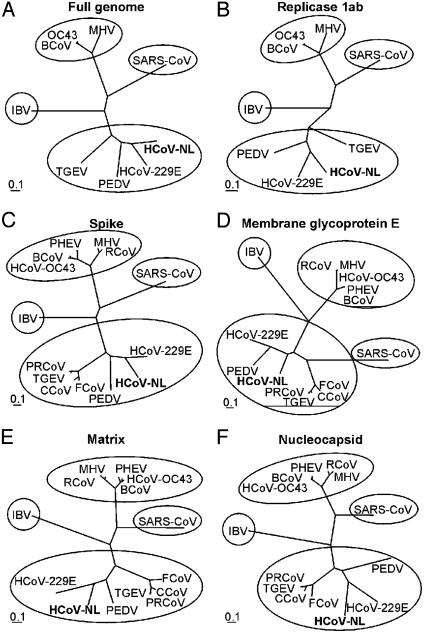
Fig. 3.
Phylogenetic analysis of HCoV-NL and other coronaviruses. The full-length genome (A), replicase 1ab (B), spike (C), membrane glycoprotein E (D), matrix protein (E), and nucleocapsid protein (F) nucleotide sequences were aligned with those of known coronaviruses and maximum likelihood trees were constructed by using 100 bootstraps and three jumbles. Bars roughly represent 10% nucleotide sequence differences between closely related strains. PEDV, porcine epidemic diarrhea virus; TGEV, transmissible gastroenteritis virus; BCoV, bovine coronavirus; MHV, murine hepatitis virus; IBV, infectious bronchitis virus; SARS-CoV, SARS coronavirus; PRCoV, porcine respiratory coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; RCoV, rat coronavirus; FCoV, feline coronavirus; CCoV, canine coronavirus.