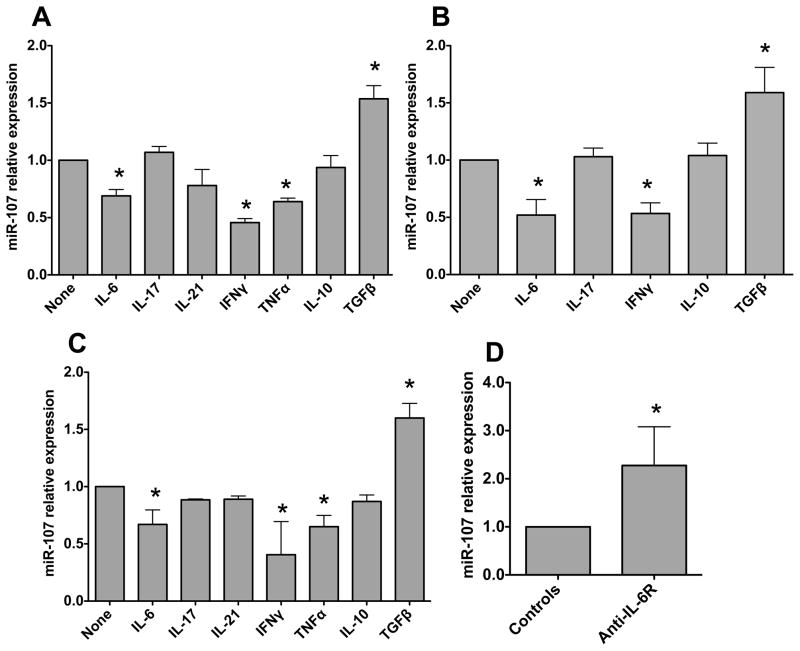
FIGURE 2. Cytokines differentially regulate DC and macrophage miR-107 expression.
(A) BMDCs were generated from C57BL/6 mice and treated with the indicated cytokines for 24 h. miR-107 expression was then analyzed by quantitative PCR, and Gapdh was used as a house-keeping gene control. miR-107 expression in BMDCs was considered baseline and arbitrarily set to 1.0, and the relative changes were calculated based on the baseline expression. (B) Human monocyte-derived DCs (MoDC) were generated from PBMCs and treated with the indicated cytokines for 24 h. miR-107 expression was analyzed by quantitative PCR with Gapdh as a house-keeping gene control. (C) BMMs were generated from C57BL/6 mice and treated with the indicated cytokines for 24 h. miR-107 expression was analyzed by quantitative PCR with Gapdh as a house-keeping gene control. Data are shown as mean ± SD of two samples from one of two experiments performed. *p<0.05 compared with controls, unpaired, two tailed Student’s t test. (D) CD4+ T cells from CBir1 Tg mice were transferred into groups of 4 RAG−/− mice and treated with anti-IL-6R antibody or control antibody from the day of cell transfer. Six weeks after transfer, the mice were sacrificed and intestinal RNA was isolated. miR-107 expression in individual mouse was determined by real-time PCR. Gapdh was used as a control. Data are shown as mean ± SD of four samples from one of two experiments performed. *p<0.05, compared with controls, unpaired, two tailed Student’s t test.