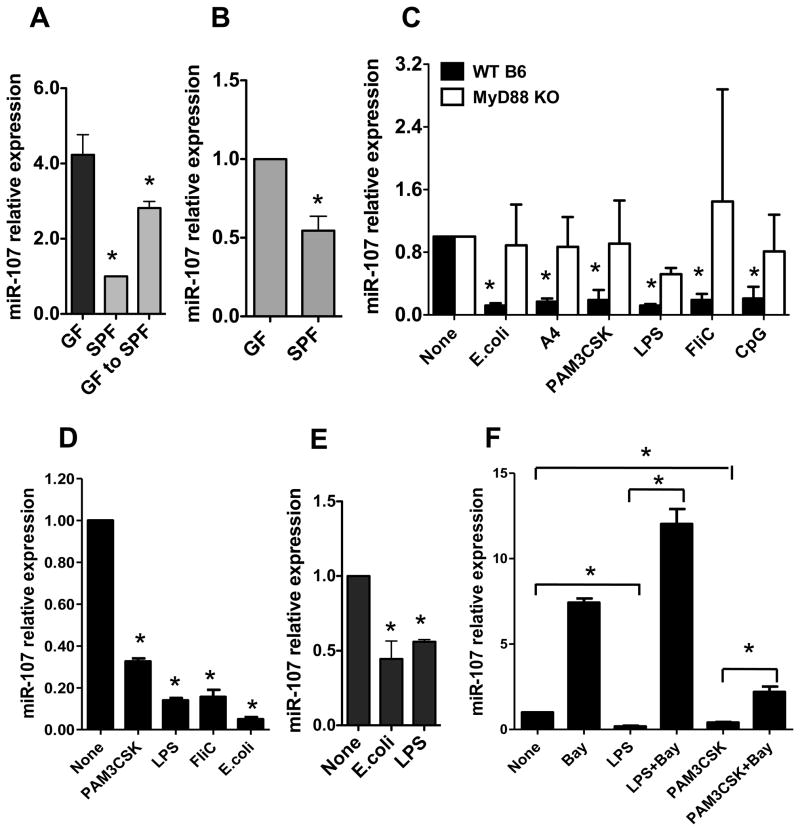
FIGURE 3. Commensal bacteria downregulate miR-107.
RNA was isolated from (A) the intestines or (B) intestinal CD11c+ cells of C57BL/6 mice housed under SPF and GF conditions, or GF C57BL/6 mice that had been recolonized with normal flora by housing under SPF conditions for 1 month. miR-107 expression was analyzed by real-time PCR. miR-107 expression in SPF mice was considered baseline and arbitrarily set to 1.0, and the relative changes were analyzed based on the baseline expression. Data are shown as mean ± SD of two samples from one of two experiments performed. *p<0.05 compared with GF mice, unpaired, two tailed Student’s t test. (C) BMDCs were generated from WT or MyD88 KO mice and treated with commensal E. coli, flagellated A4 bacteria or various TLR ligands for 24 h. miR-107 expression was analyzed by real-time PCR. The miR-107 expression in BMDCs was considered baseline and arbitrarily set to 1.0, and the relative changes were analyzed compared with the baseline expression. Data are shown as mean ± SD of two samples from one of three experiments performed. (D) Human monocyte-derived DCs were stimulated with TLR ligands and miR-107 expression was analyzed by real-time PCR. Data are shown as mean ± SD of two samples from one of three experiments performed. (E) BMMs were stimulated with E. coli or LPS, and miR-107 expression was analyzed by real-time PCR. Data are shown as mean ± SD of two samples from one of two experiments performed. (F) BMDCs were treated with TLR ligands with or without NF-κB inhibitor (Bay 11–7082) for 24 h. miR-107 expression was analyzed by quantitative PCR. Data are shown as mean ± SD of two samples from one of three experiments performed. (C–F) *p<0.05 compared with controls, unpaired, two tailed Student’s t test.