Summary
In this study we examined the role IL-13 receptor alpha 1 (IL-13Rα1) plays in macrophage differentiation and function. The findings indicate that IL-13Rα1 is expressed on the M2 but not the M1 subset of macrophages and specifically heterodimerizes with the IL-4Rα chain to form a type II receptor, which controls the differentiation and function of these cells. Indeed, bone marrow (BM) cells from IL-13Rα1+/+ and IL-13Rα1−/− mice yield equivalent numbers of macrophages when cultured under M2 polarizing conditions. However, IL-13Rα1−/− BM cells yield a much higher number of macrophages than IL-13Rα1+/+ BM cells when the differentiation is carried out under M1-polarizing conditions. Further analyses indicated that macrophages that express IL-13Rα1 also display surface markers associated with an M2 phenotype. In addition, the IL-13Rα1+ macrophages were highly efficient in phagocytizing zymosan bioparticles both in vitro and in vivo, and supported differentiation of naïve T cells to a Th2 phenotype. Finally, when stimulated by IL-13, a cytokine that uses the heteroreceptor, the cells were able to phosphorylate STAT6 efficiently. These previously unrecognized findings indicate that IL-13Rα1 serves as a marker for M2 macrophages and the resulting heteroreceptor influences both their differentiation and function.
Keywords: Antigen presentation, Differentiation, IL-13 Rα1, macrophages, Phagocytosis
Introduction
Macrophages have been classified into inflammatory M1 and anti-inflammatory M2 subsets on the basis of the effector function associated with their cytokines [1–3]. Specifically, M1 macrophages produce the pro-inflammatory cytokines IL-6, IL-12 and TNF-α while M2 macrophages secrete IL-10 [4–7]. In addition, M1 macrophages produce nitric oxide while M2 express arginase 1 (Arg-1) [3–5, 8]. Later, surface markers were identified that were associated with either M1 or M2 macrophage subsets and these include FIZZ1, Ym1 and Ly6C [9, 10]. Also, expression of receptors such as CCR4 chemokine receptor [11], IL-12 receptor beta 2 (IL-12Rβ2) [12], IL-4 receptor alpha (IL-4Rα) [13, 14], the mannose receptor (MR), CD206 [15], and IL-21 receptor [16], is subset specific and likely shapes the function of the different macrophage populations. Furthermore, it has been reported that signaling molecules such as STAT6 [13] and Akt1 and 2 [17] contribute to the polarization of macrophages into their subsets. Recent studies have reported that mice deficient for IL-4Rα, in which both the heteroreceptor (IL-4Rα/IL-13Rα1) and the conventional IL-4R (IL-4Rα/γ) are non-functional, manifest a bias towards M1 phenotype [18]. Also, mice deficient for IL-13, which usually signal through IL-4Rα/IL-13Rα1 heteroreceptor, display a bias towards the M1 macrophage phenotype [19]. Even in human cells, knock down of IL-13Rα1 with micro RNA converts macrophages from M2 to M1 phenotype [20]. These observations suggest that the IL-4Rα/IL-13Rα1 heteroreceptor plays a role in macrophage phenotype and function. Herein, we used IL-13Rα1-deficient mice [21], in which the heteroreceptor (IL-4Rα/IL-13Rα1) is non-functional but the conventional IL-4R (IL-4Rα/γ) is intact, to determine whether this heteroreceptor influences macrophage differentiation and function. Furthermore, we have taken advantage of the availability of IL-13Rα1-GFP reporter mice and the anti-IL-13Rα1 monoclonal antibody (both of which were produced in our laboratory) [21] to gain further insight into how the heteroreceptor regulates macrophage development and function. To our surprise, we found that IL-13Rα1 expression serves as a surface marker for M2 macrophages and distinguishes this population from the M1 subset. In addition, we demonstrated that IL-13Rα1 on the surface of M2 macrophage is able to form a heterodimer with IL-4Rα and the resulting heteroreceptor sustains a strong STAT6 phosphorylation upon stimulation with IL-13, an indication of receptor functionality on M2 macrophages. In fact, receptor signaling likely contributed to the functional characteristics of M2 macrophages, including morphological changes and adaptation to different tissues.
Results
IL-13Rα1 is predominantly expressed on dendritic cells and monocytes in both adult and neonatal mice
The contribution of IL-13Rα1 to the development and maturation of cells of the immune system, as well as its involvement in immune responses, remains largely undefined most likely due to the lack of reagents that detect and/or interfere with the function of the receptor. Recently, we generated a mouse strain in which the IL-13Rα1 gene was disrupted [21] and used these animals for immunization with recombinant IL-13Rα1 protein to generate a monoclonal antibody against the receptor. One hybrid clone designated as 1G3 produces a monoclonal anti-IL-13Rα1 antibody that specifically binds to CTLL-2 cells transfected with IL-13Rα1vector, but not to non-transfected cells (Supporting Information Fig. 1).
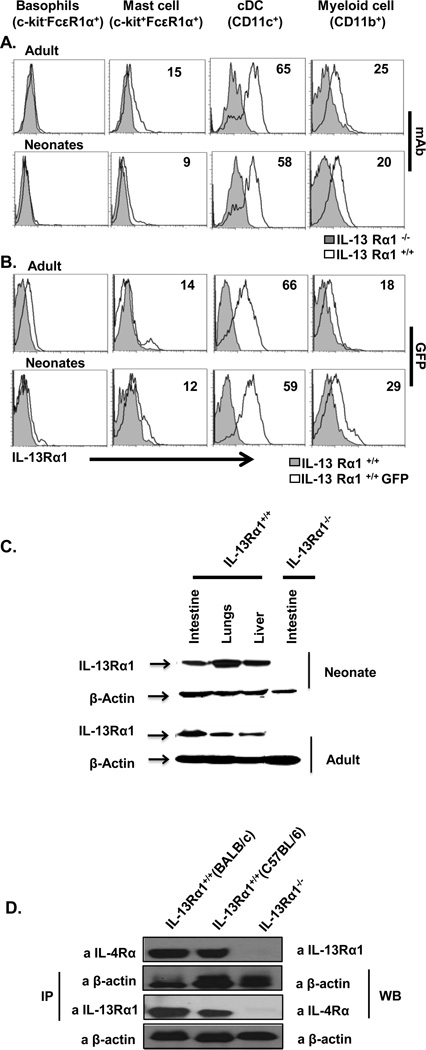
Previous studies have shown that IL-13Rα1 is variably expressed in adult and neonatal mice thereby implicating this receptor in the functional discrepancies of the developing immune system [22, 23] and among multiple cell types [21, 24]. Therefore, we analyzed receptor expression on cells of the innate and adaptive immune system, in both adult and neonatal mice. The findings show that cells of the adaptive immune system do not bind anti-IL-13Rα1 antibody in either neonatal or adult mice, which indicates that the receptor is not expressed on these cells (Supporting Information Fig. 2). However, both CD11c and CD11b cells display receptor expression in both the neonatal and adult mice, as detected by anti-IL-13Rα1 antibody (Fig.1A). Moreover, the findings corroborate with studies that used IL-13Rα1-GFP reporter mice, where GFP (IL-13Rα1) expression was evident in CD11c and CD11b cells, but not in other cells, whether in neonatal or adult mice (Fig. 1B). Overall, IL-13Rα1 is mostly expressed on CD11c and CD11b cells which are mostly conventional DCs and myeloid cells.
Figure 1.
IL-13Rα1 is expressed on various cell types and forms a heteroreceptor with the IL-4Rα chain. Splenic cells from IL-13Rα1+/+, IL-13Rα1−/− and IL-13Rα1-GFP reporter neonatal and adult C57BL/6 mice were stained for basophil, mast cell, dendritic cell, and macrophage markers. (A) Cells from IL-13Rα1+/+ and IL-13Rα1−/− mice were also stained with an anti-IL-13Rα1 monoclonal antibody and 7AAD, and IL-13Rα1 expression was analyzed on live cells (7AAD−), gated on the indicated cell type. (B) Splenic cells from IL-13Rα1-GFP reporter neonatal and adult C57BL/6 mice were stained with 7AAD but not with anti-IL-13Rα1 monoclonal antibody and IL-13Rα1 expression was determined by analysis of GFP expression on live cells (7AAD−) gated on the indicated cell type. Numbers represent the percentage of cells expressing IL-13Rα1. Results obtained with cells pooled from 2 mice are shown and are representative of 4 independent experiments. (C) Expression of IL-13Rα1 in intestinal, lung and liver cells of neonatal and adult IL-13Rα1+/+ BALB/c mice as determined by western blot using an anti-IL-13Rα1 mAb. Intestinal epithelial cells from adult and neonatal IL-13Rα1−/− mice were included as a negative control. Results show samples from 2 mice and the data are representative of 3 independent experiments. (D) Immunoprecipitation (IP) of the heteroreceptor by anti-IL-4Rα and western blotting (WB) of IL-13Rα1 with anti-IL-13Rα1 Ab (top). For control purposes, equivalent amounts of lysate were subjected to IP with anti-β-actin antibody and blotted with the same antibody. In the bottom panel, the heteroreceptor immunoprecipitated with anti-IL-13Rα1 and IL-4Rα was blotted with anti-IL-4Rα Ab. (B, C) β-actin was used as a loading control. Intestinal cells from IL-13Rα1−/− BALB/c mice were included as negative control. The data are representative of three independent experiments obtained with cells pooled from 2 mice per experiment.
The IL-13Rα1 chain binds IL-13 with very low affinity [25]. However, it was suggested that IL-13Rα1 might associate with the IL-4Rα chain to form a heteroreceptor through which both IL-4 and IL-13 can signal [25–27]. Using our new monoclonal antibody we were able to show that cells in different organs from both adult and neonatal mice expressed IL-13Rα1 and this chain co-precipitated with IL-4Rα (Fig 1). Indeed, cell lysates from the intestine, lung and liver of both neonatal and adult IL-13Rα1+/+ BALB/c mice stained positive with anti-IL-13Rα1 monoclonal antibody 1G3 as analyzed by western blot (Fig. 1C). Moreover, when IL-13Rα1 was precipitated with 1G3 antibody it was found to co-precipitate with IL-4Rα, as is shown by the 140 kDa band detected with anti-IL-4Rα antibody (Fig. 1D). Similarly, precipitation of IL-4Rα with anti-IL-4Rα antibody allowed co-precipitation of a 65 kDa protein that was detected with 1G3 mAb (Fig.1D). Overall, IL-13Rα1 is expressed by many cell types in both adults and neonates, and forms a heteroreceptor with IL-4Rα.
IL-13Rα1 expression on macrophages is associated with the M2, but not M1 phenotype
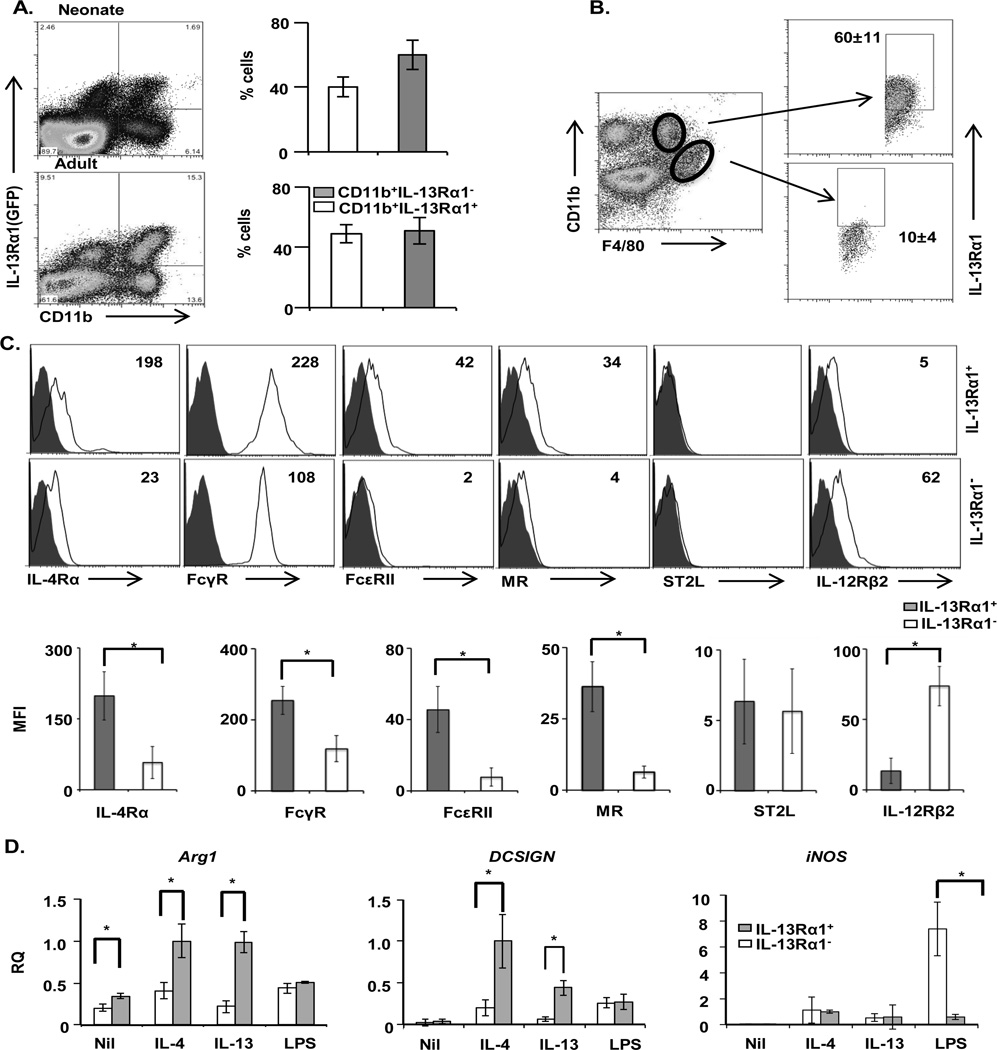
The conventional IL-13R, which is formed by a complex of IL-13Rα1 and IL-13Rα2 subunits, is generally thought of as a decoy receptor, which may act to sequester circulating IL-13 cytokine [28, 29]. On the other hand, it was suggested that much of the signaling function of IL-13 is carried out through the IL-13Rα1/IL-4Rα type II heteroreceptor [30]. Since this heteroreceptor is mainly expressed on CD11b+ cells, a population that is rather diverse in phenotype and function, we sought to investigate the role IL-13Rα1 plays on the phenotypic and functional assortment of these cells. To this end, spleen cells from IL-13Rα1-GFP reporter mice were stained with anti-CD11b antibody, and IL-13Rα1 expression was analyzed on CD11b-gated cells. Interestingly, the results show a bimodal expression of IL-13Rα1 among CD11b+ cells from adult and neonatal mice (Fig. 2A). In fact, among neonatal CD11b+ cells, IL-13Rα1 was expressed on 40% of the population, while in adults, receptor expression was displayed on about 50% of the cells. Thus, in neonates as well as adult animals about half of CD11b+ cells express the receptor while the other half do not. To determine whether IL-13Rα1 expression is associated with a particular Macrophage phenotype, the CD11b+ splenic cells were stained for F4/80, a marker that distinguishes Macrophage phenotype on the basis of anatomic location, and IL-13Rα1 expression was analyzed on CD11bhiF4/80lo white pulp- and CD11bloF4/80hi red pulp-splenic macrophages. The results clearly show that IL-13Rα1 is expressed on the CD11bhiF4/80lo white pulp macrophages (Fig. 2B). Besides distinction on the basis of localization, macrophages can be further divided into M1 and M2 on the basis of marker expression such as IL-4Rα, FcγR, FcεRII, mannose receptor (MR), IL-33 receptor (ST2L) and IL-12Rβ2 [1–3]. We then tested CD11b+ F4/80+IL-13Rα1+ macrophages for the expression of these markers in comparison with IL-13Rα1− (CD11b+ F4/80+IL-13Rα1−) cells. The results show that CD11b+F4/80+IL-13Rα1+ macrophages have higher expression of IL-4Rα (MFI of 198 vs. 23), FcγRII/III (MFI of 228 vs. 108), FcεRII, (MFI of 42 vs. 2) and MR (34 vs. 4) relative to CD11b+F4/80+IL-13Rα1− macrophages (Fig. 2C). Compiled results from 3 experiments showed that the results are statistically significant and confirmed that IL-13Rα1 expression is associated with the M2 phenotype of macrophages. Moreover, expression of IL-12Rβ2, which is usually associated with the M1 phenotype, is expressed at a much higher level on the CD11b+F4/80+IL-13Rα1− population, relative to CD11b+F4/80+IL-13Rα1+ macrophages (MFI of 62 vs. 5) (Fig. 2C). ST2L (IL-33Rα) expression, which is associated with the M2b Macrophage subtype [31], was not detected on either type of macrophages (Fig. 2C).
Figure 2.
IL-13Rα1 expression is associated with M2 Macrophage phenotype. (A) Splenocytes pooled from 3 adult or neonatal IL-13Rα1+/+-GFP mice were stained with anti-CD11b antibody and IL-13Rα1 expression on CD11b+ cells was analyzed by flow cytometry. The bar graphs show the mean percentage of CD11b+ cells ± SD from 3 independent experiments. (B) Splenocytes from individual adult IL-13Rα1+/+-GFP mice were stained with anti-CD11b and anti-F4/80 antibodies and the different subsets of macrophages were identified by flow cytometry, on the basis of CD11b and F4/80 expression (left). The right panel shows GFP (IL-13Rα1) expression on CD11bhiF4/80Lo and CD11bLoF4/80hi macrophages. The numbers indicate the mean percent ± SD of IL-13Rα1 expression compiled from five individual mice. Data are representative of 3 independent experiments. (C) Splenocytes pooled from 2 IL-13Rα1+/+ adult C57BL/6 mice were stained for CD11b, F4/80, IL-4Rα, FcγR (II/III), FcεRII, mannose receptor (MR), ST2L (IL-33R), and IL-12Rβ2 and analyzed by flow cytometry. The cells gated on CD11b and F4/80 were analyzed for expression of the indicated markers (open histograms) and plotted relative to the corresponding isotype (filled histograms). The numbers in the upper right corners indicate the MFI for each marker. Bar graphs in the lower panel show the mean ± SD of MFI for the indicated markers compiled from 4 experiments. (D) Purified splenic CD11b+F4/80+IL-13Rα1+ and CD11b+F4/80+IL-13Rα1− macrophages (5 × 104 cells/well) were stimulated with IL-4, IL-13, LPS or media alone (Nil) for 4 h and total RNA was extracted using Trizol. The mRNA levels for Arg1, murine DCSIGN (SIGNR3), and iNOS were determined by RT-qPCR as described in material and methods. Each bar represents the mean fold change ± SD of triplicate samples in one of 3 representative experiments. *p < 0.05 (unpaired two-tailed student t test)
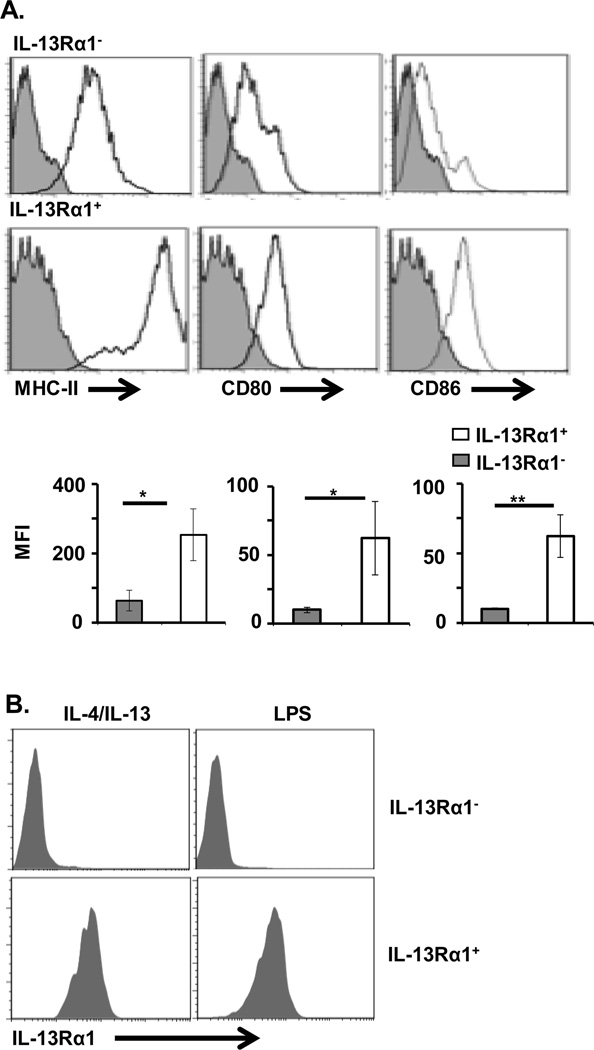
Given that M2, but not M1, macrophages express IL-13Rα1, and that this chain associates with IL-4Rα to form the type II heteroreceptor through which both IL-4 and IL-13 can signal, we sought to determine whether M2 macrophages utilize the heteroreceptor to carry out functions usually associated with their phenotype. Accordingly, CD11b+F4/80+IL-13Rα1+ splenic cells were isolated by FACS and briefly stimulated with either IL-4 or IL-13 cytokines. Subsequently, the levels of Arg1 and DCSIGN (SIGNR3) transcripts, which are known to be elevated in functional M2 macrophages [32], were assessed by RT-qPCR. The results showed that both Arg1 and DCSIGN transcripts are significantly increased in CD11b+F4/80+IL-13Rα1+, relative to CD11b+F4/80+IL-13Rα1− macrophages (Fig. 2D). In contrast, LPS, which is known to stimulate inducible nitric oxide synthase (iNOS) expression in M1 macrophages [33], significantly increases iNOS expression in CD11b+F4/80+IL-13Rα1− (M1) macrophages, relative to CD11b+F4/80+IL-13Rα1+ (M2) macrophages (Fig. 2D). Furthermore, analysis of MHC-II and costimulatory molecule expression showed that IL-13Rα1+ macrophages have a pattern of MHC-II and co-stimulatory molecule expression typical of M2 macrophages, while IL-13Rα1− cells express these molecules at lower levels reminiscent of M1 macrophages (Fig. 3A). These findings suggest that IL-13Rα1+ cells would be efficient in phagocytosis and Ag presentation, as is the case for M2 macrophages [2, 34]. In addition, IL-13Rα1− macrophages emanating from IL-13Rα1+/+ mice that possess the potential for receptor up-regulation remained IL-13Rα1-negative upon stimulation with LPS or IL-4/IL-13 (Fig. 3B). However, IL-13Rα1+ macrophages remained IL-13Rα1-positive under either stimulation condition (Fig. 3B). Thus, expression, or the lack thereof, of IL-13Rα1 upon Macrophage phenotype commitment is stable, suggesting that the receptor serves as a reliable marker for the subset. Overall, IL-13Rα1+ macrophages display a gene and surface expression profile typically associated with M2 macrophages.
Figure 3.
IL-13Rα1 is stably expressed on macrophages that display high levels of MHC and costimulatory molecules. Purified splenic CD11b+F4/80+IL-13Rα1+ and CD11b+F4/80+IL-13Rα1− macrophages from IL-13Rα1+/+-GFP mice were stained for MHC II and costimulatory molecules and analyzed by flow cytometry. (A) Expression of MHC II, CD80 and CD86 on IL-13Rα1+ and IL-13Rα1− Macrophage populations. The upper panel shows representative flow cytometry data from 5 experiments, while the bottom panel shows mean ± SD of MFI data compiled from 5 experiments. *p < 0.05, ** p< 0.005 (unpaired two-tailed student t test). (B) IL-13Rα1+ and IL-13Rα1− Macrophage populations were stimulated with a mixture of IL-4 and IL-13 (30 ng/mL of each cytokine), or LPS (100 ng/mL) for 24 h and GFP (IL-13Rα1) expression was analyzed by flow cytometry.
IL-13Rα1+ macrophages display functional traits associated with M2 type macrophages
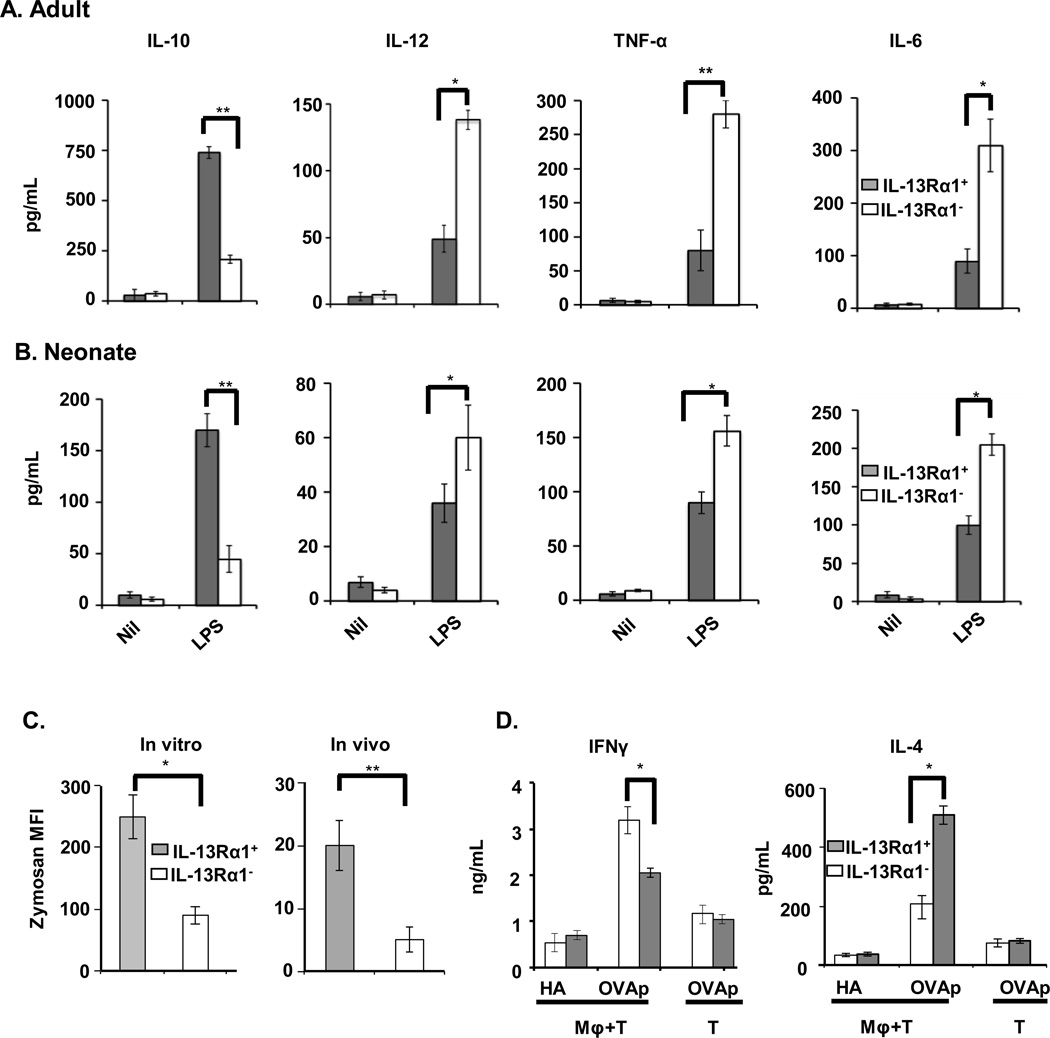
Since M1 macrophages are generally regarded as inflammatory, while M2 macrophages are considered anti-inflammatory, we tested both IL-13Rα1+ (M2 phenotype) and IL-13Rα1− (M1 phenotype) subsets for production of inflammatory cytokines and for the ability to carry out phagocytic function. The results indicated that both neonatal and adult CD11b+F4/80+IL-13Rα1+ macrophages produce significant amounts of the anti-inflammatory cytokine IL-10, but lower levels of the pro-inflammatory cytokines IL-12, TNFα and IL-6 upon stimulation with LPS as compared with control media (Fig. 4A and B). In contrast, CD11b+F4/80+IL-13Rα1− macrophages produce higher levels of the pro-inflammatory cytokines IL-12, TNFα, and IL-6, but very little IL-10 (Fig. 4A and B). Moreover, since IL-13Rα1+ macrophages express significant levels of the mannose receptor (MR), we envisioned that they would be highly phagocytic as has been previously shown for M2 macrophages [35]. To test this premise we sorted CD11b+F4/80+IL-13Rα1+ and CD11b+F4/80+IL-13Rα1− macrophages, and evaluated their ability to phagocytize opsonin-coated zymosan bio-particles. The results showed that the phagocytic activity of IL-13Rα1+ macrophages is significantly higher than that of the IL-13Rα1− macrophages, both in vitro and in vivo (Fig. 4C). Indeed, the MFI observed with IL-13Rα1+ macrophages is 2.5-fold higher than the MFI obtained with IL-13Rα1− macrophages, indicating that the former are much more effective at ingesting the zymosan bio-particle in vitro (Fig. 4C, left panel). Similarly, the IL-13Rα1+ macrophages ingested threefold more Texas Red zymosan bio-particles than their IL-13Rα1− counterparts in vivo (Fig. 4C, right panel). In addition, since M1 macrophages produce IL-12 and polarize naïve T cells towards Th1, while M2 macrophages support development of Th2 cells [5], we tested both IL-13Rα1+ and IL-13Rα1− macrophages for their ability to stimulate naïve T cells towards differentiation along the Th1 and Th2 pathways. To this end, OVA323-329-specific TCR transgenic OT-II CD4+ T cells [36] were cultured with CD11b+F4/80+IL-13Rα1+ or CD11b+F4/80+IL-13Rα1− macrophages in the presence of OVA323-329 peptide, and the production of IL-4 and IFN-γ was measured by ELISA. The results show that IL-13Rα1+ macrophages induce Th2-cell differentiation, while IL-13Rα1− macrophages support Th1-cell differentiation (Fig. 4D). Indeed, in the culture with IL-13Rα1+ macrophages there was significant production of IL-4, relative to OVAp-loaded IL-13Rα1− macrophages. Also, IL-4 production was much higher than with IL-13Rα1+ cells loaded with the control hemagglutinin (HA) peptide, or with T cells stimulated with OVAp without any macrophages (Fig. 4D, right panel). In contrast, co-culture experiments produced significantly higher IFNγ when OVAp was presented by IL-13Rα1− versus IL-13Rα1+ macrophages (Fig. 4D, left panel). Overall, IL-13Rα1+ macrophages produce IL-10, display efficient phagocytic functions, and polarize T cells towards Th2 cells as usually observed with M2 macrophages.
Figure 4.
IL-13Rα1+ macrophages are highly phagocytic and produce mostly IL-10. Purified CD11b+F4/80+IL-13Rα1+ and CD11b+F4/80+IL-13Rα1− macrophages (2 × 105/well) from (A) two adult and (B) six neonatal mice were stimulated with LPS (100 ng/mL) or media alone (Nil) for 24 h and the amount of IL-6, IL-10, IL-12, and TNF-α in the supernatants was measured by ELISA. Bar graphs show mean ± SD of triplicate wells from 3 independent experiments. (C) Zymosan uptake by macrophages in vitro is shown in the left panel. Texas Red labeled zymosan bio-particles (1 × 104/well) were incubated with CD11b+F4/80+IL-13Rα1+ or CD11b+F4/80+IL-13Rα1− macrophages (1 × 105/well) for 3 h and ingested particles were measured by flow cytometry. The right panel shows in vivo zymosan uptake. Texas Red labeled zymosan bio-particles were injected i.v. into IL-13Rα1-GFP C57BL/6 mice (1 × 104 bio-particle per/mouse) and spleen cells were harvested 24 h later. Cells were stained with anti-CD11b and F4/80 antibodies and the CD11b+F4/80+GFP+ (IL-13Rα1+) and CD11b+F4/80+GFP− (IL-13Rα1−) cells were analyzed for zymosan bio-particles by flow cytometry. Data is shown as mean ± SD of MFI from 3 samples in 3 independent experiments. (D) Naïve OVAp specific TCR transgenic OT-II CD4+ T cells (3 × 105 cells/well) were stimulated with irradiated IL-13Rα1+ or IL-13Rα1− macrophages (3 ×104/well) (Macrophage +T) in presence of OVA or HA peptide and IL-4 or IFNγ in the culture supernatant were measured by ELISA. OT-II CD4+ T cells incubated with OVAp without macrophages (T) were included as a control. The bar graphs show the mean ± SD of triplicate wells and are representative of 3 independent experiments. *p < 0.05, ** p <0.005 (unpaired two-tailed student t test).
IL-13Rα1 expression influences macrophage differentiation
The association of IL-13Rα1 expression with the M2 macrophage phenotype raises the question as to whether receptor expression influences the differentiation of BM myeloid progenitors into M2 macrophages. It is well known that GM-CSF and M-CSF promote macrophage differentiation into M1 and M2 phenotypes, respectively [37]. We used these cytokines to differentiate BM cells isolated from IL-13Rα1+/+ and IL-13Rα1−/− mice into macrophages to test whether IL-13Rα1 contributes to lineage commitment. The results show that BM cells from IL-13Rα1+/+ and IL-13Rα1−/− mice cultured with M-CSF yield about equivalent numbers of CD11b+F4/80+ macrophages (Fig. 5A). Among these macrophages, both MR and IL-4Rα are expressed at a similar level, suggesting that IL-13Rα1 does not play a significant role in M2 differentiation (Fig. 5A). However, when BM cells were stimulated with GM-CSF, there was a higher frequency of CD11b+F4/80+ macrophages among IL-13Rα1− cultures, compared with IL-13Rα1+/+ BM donors (Fig. 5B). Moreover, when the macrophages were analyzed for phenotypic markers, BM cells from IL-13Rα1−/− mice cultured with GM-CSF, which drives M1 differentiation, had much higher expression of IL-1R and IL-12Rβ2 (M1 phenotype), compared with IL-13Rα1+/+ mice suggesting that IL-13Rα1 expression influences the differentiation of M1 macrophages (Fig. 5B). Since there was no differential expression of MCSF-1R or GM-CSFRα among IL-13Rα+ and IL-13Rα1− granulocyte-macrophage progenitor (GMP), it is likely that macrophage differentiation into phenotypic subsets is influenced by IL-13Rα1 expression (Supporting Information Fig. 3). In addition, iNOS transcription is observed in GM-CSF/IL-13Rα1−/− BM culture and is unaffected by IL-13, perhaps due to the lack of functional IL-13Rα1/IL-4Rα heteroreceptor. However, in the presence of IL-4, iNOS transcription was reduced to a minimal level, as the conventional IL-4R remains intact in these cells (Fig. 5C). Furthermore, in the GM-CSF/ IL-13Rα1+/+ BM culture, iNOS gene expression was much lower in comparison to the IL-13Rα1−/− BM culture. These results suggest that IL-13Rα1 influences both differentiation and function of M1 macrophages. Similarly, the GM-CSF differentiated culture from IL-13Rα1−/− BM cells had increased M1-associated cytokines such as IL-12, TNFα and IL-6 upon stimulation with LPS, while IL-10 was equivalent in both IL-13Rα1−/− and IL-13Rα1+/+ cultures (Fig. 5D). Again, these data implicate IL-13Rα1 in the differentiation and function of M1 macrophages, especially since M-CSF/IL-13Rα1−/− cultures stimulated with LPS do not produce an altered cytokine profile, compared with IL-13Rα1+/+ derived cultures (Fig. 5D). Overall, IL-13Rα1 influences macrophage differentiation by limiting the generation of M1 macrophages.
Figure 5.
Macrophage differentiation shifts towards an M1 phenotype in the absence of IL-13Rα1. Bone marrow (BM) stem cells were harvested from adult IL-13Rα1−/− or IL-13Rα1+/+ (closed bars) mice and cultured in presence of the growth factor GM-CSF or M-CSF for 7 days. (A) The M-CSF culture was stained with anti-CD11b, anti-F4/80, anti-IL-4Rα, and anti-MR. The total number of CD11b+F4/80+ (left panel) as well as the number of CD11b+F4/80+ macrophages expressing M2 (IL-4Rα+ and MR+) phenotype (right panel) are shown. (B) The GM-CSF culture was stained with anti- CD11b, anti-F4/80, anti-IL-1R, and anti-IL-12Rβ2 antibodies. The total number of CD11b+F4/80+ macrophages (left panel) as well as the number of CD11b+F4/80+ macrophages expressing M1 (IL-1R+ and IL-12Rβ2+) phenotype (right panel) are shown. The symbols represent individual mice and the bars represent mean ± SD of 4 samples compiled from three independent experiments. (C) BM stem cells were cultured with GM-CSF in the presence of media alone (Nil) or media supplemented with either IL-4 or IL-13. After 7 days the cells were lysed, RNA was extracted and used to measure iNOS expression by quantitative RT-PCR. The bars show relative coefficient (RQ) of gene expression using GAPDH as baseline. The data are presented as mean ± SD of 3 samples from 3 independent experiments. (D) GM-CSF- or M-CSF-derived CD11b+ macrophages (5 × 105) were purified, stimulated with LPS (100 ng/mL) or media alone (Nil) and cytokine production in the culture supernatant was measured by ELISA. The bar graphs show the mean ± SD of triplicate wells from one of 3 representative experiments. *p < 0.05, **p < 0.001 (unpaired two tailed student t test).
IL-13 induces STAT6 phosphorylation and unique morphological changes in IL-13Rα1+/+ macrophages
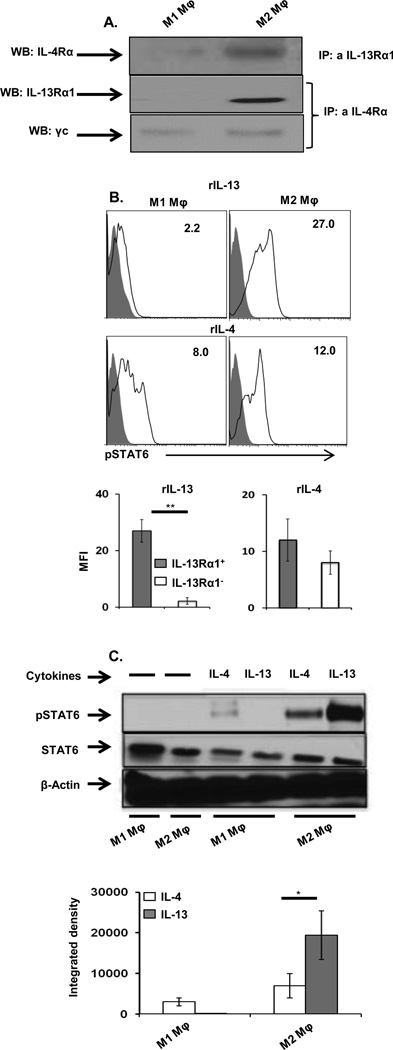
IL-13Rα1 is expressed on M2 macrophages and likely forms a heteroreceptor by association with IL-4Rα, through which both IL-4 and IL-13 signal and trigger activation and function. To test these premises, we began by assessing expression of the IL-4Rα/IL-13Rα1 heteroreceptor, as well as conventional IL-4R (IL-4Rα/γ) on M2 macrophages. The results in Figure 6A show that immunoprecipitation of IL-13Rα1 with anti-IL-13Rα1 antibody pulled down IL-4Rα from IL-13Rα1+ M2 macrophages, but not IL-13Rα1− macrophages, indicating that the former express IL-4Rα/IL-13Rα1 heteroreceptor. Similarly immunoprecipitation of IL-4Rα with anti-IL-4Rα antibody pulled down IL-13Rα1 from IL-13Rα1+ M2 macrophages, but not IL-13Rα1− macrophages, providing confirmation that M2 macrophages express the heteroreceptor. The fact that IL-4Rα precipitation pulled down the internal control common γ chain in both IL-13Rα1+ and IL-13Rα1− macrophages, indicates that the differential precipitations are not related to inaccurate sample loading (Fig. 6A). These results indicate that M1 macrophages express the conventional receptor (IL-4Rα/γ) while M2 macrophages express both receptors (conventional IL-4Rα/γ and heteroreceptor IL-4Rα/IL-13Rα1).
Figure 6.
STAT6 phosphorylation is greater in IL-13Rα1+ than in IL-13Rα1− (A) Purified CD11b+F4/80+IL-13Rα1− (M1 macrophages) and CD11b+F4/80+IL-13Rα1+ (M2 macrophages) macrophages were lysed and immunoprecipitation of IL-4Rα by anti- IL-13Rα1 antibody was performed (top). The middle and bottom lanes show immunoprecipitation of IL-13Rα1 and the common gamma chain (γc) by anti-IL-4Rα antibody, respectively. Experiment was repeated 2 times with similar results. . (B) Purified M1 and M2 Macrophage (5 × 104/well) were stimulated with recombinant murine IL-4 or IL-13 (30 ng/mL) or medium alone for 30 minutes and phosphorylation of STAT6 was determined by (B) intracellular staining with an anti-pY641STAT6 antibody for cytokine stimulated cells (open histogram) compared with to cells that were cultured with media alone (closed histogram). The number in the upper right corner indicates the MFI of phosphorylated STAT6. The bar graphs show mean ± SEM of compiled MFI results from 3 samples in 3 independent experiments. (C) Purified M1 and M2 Macrophage were stimulated as in (B) and STAT6 phosphorylation was determined by western blot. The top panel shows a representative blot depicting phosphorylated (top) and unphosphorylated (middle) STAT6 as well as the control β-actin (bottom). The bottom panel shows the mean ± SD of integrated density for the phosphorylated STAT6 bands using data from 3 samples. The results are representative of 2 independent experiments. *p < 0.05, **p < 0.005 (unpaired two-tailed student t test).
The heteroreceptor on IL-13Rα1+ M2 macrophages is functional, because IL-13 induces significant phosphorylation of STAT6. Indeed, incubation with IL-13, which binds only the heteroreceptor, induces significant STAT6 (Y641) phosphorylation in IL-13Rα1+ M2 macrophages (Fig. 6B). No significant STAT6 phosphorylation was observed with IL-13Rα1− macrophages, which express the conventional IL-4 receptor, but not the heteroreceptor (Fig. 6B, lower panel). Incubation with IL-4 induced STAT6 (Y641) phosphorylation in both IL-13Rα1+ and IL-13Rα1− macrophages, as both cells express the conventional IL-4 receptor, but was slightly higher in IL-13Rα1+ macrophages (Fig. 6C). The greatest effect on STAT6 phosphorylation was seen in IL-13Rα1+ macrophages upon addition of IL-13 as compared with IL-13Rα1− macrophages, which did not phosphorylate STAT6. When the heteroreceptor is present, greater STAT6 phosphorylation is achieved, as IL-13 induces much higher STAT6 phosphorylation in IL-13Rα1+ macrophages than IL-4 despite the fact that unphosophorylated STAT6 levels are similar with both cytokines (Fig. 6C). This is supported by the observation that IL-4 stimulation reduces pSTAT6 in IL-13Rα1− macrophages, which express the conventional IL-4R only. The bar graphs showing integrated density compiled from 2 different experiments confirm that the stronger IL-13-induced STAT6 phosphorylation in M2 macrophages is related to heteroreceptor expression (Fig. 6C, lower panel). Overall, these results demonstrate functional IL-13 signaling through the heteroreceptor in IL-13Rα1+ macrophages, which more efficiently phosphorylates STAT6 than signaling through the conventional IL-4 receptor.
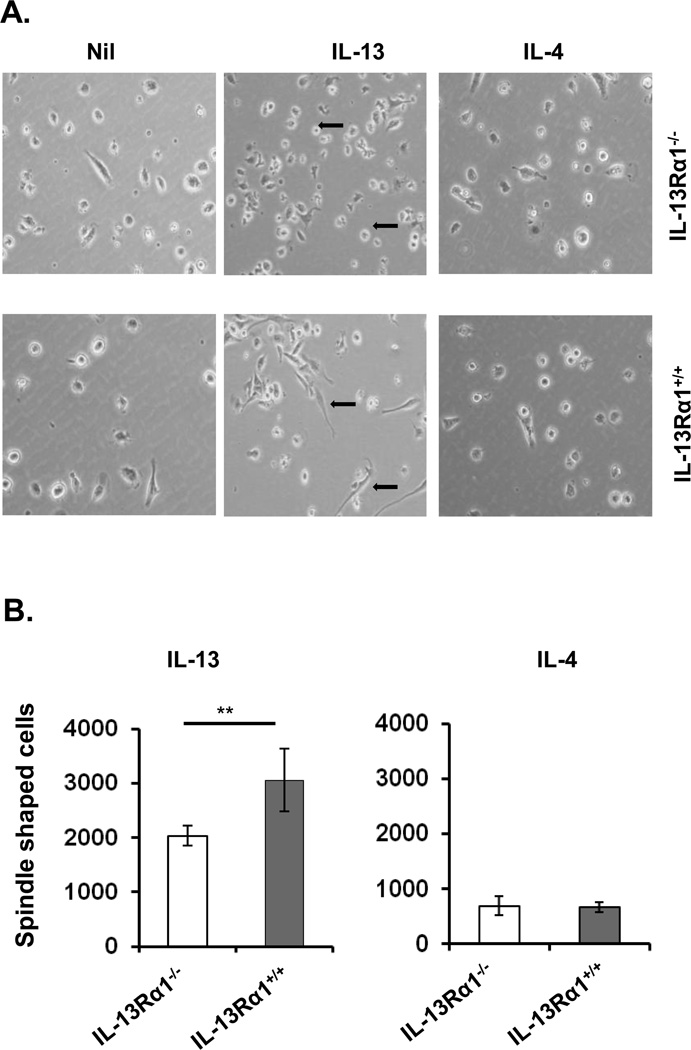
This signaling pathway also plays a role in the morphological shaping of macrophages during differentiation. Indeed, in the presence of IL-13, GM-CSF-driven differentiation of macrophages displayed a significantly increased number of fibroblast-like or spindle-shaped macrophages when the BM cells were derived from IL-13Rα1+/+ relative to IL-13Rα1−/− mice (Fig. 7). However, in the presence of IL-4, the number of spindle-shaped macrophages were at background level whether the BM cells were from IL-13Rα1+/+ or IL-13Rα1−/− mice (Fig. 7). Thus, IL-13 signaling through the heteroreceptor likely influences GM-CSF-driven macrophage differentiation.
Figure 7.
IL-13 induces unique morphologic changes in IL-13Rα1+ macrophages. Bone marrow (BM) cells from adult IL-13Rα1+/+ and IL-13Rα1−/− mice were incubated with GM-CSF for 3 days to drive maturation into macrophages. The cells were then split into 3 fractions, one of which was supplemented with media alone (Nil), the second with 30 ng/mL IL-13, and the third with 30 ng/mL IL-4. Cell morphology was analyzed by inverted light microscopy 4 days after the addition of cytokines. (A) Shows photomicrographs taken at the end of cultures. Arrows indicate morphological changes in macrophages. (B) Spindle shaped cells per 104 macrophages per well were counted and the bars represent the mean ± SD of triplicate wells. The data are representative of 3 independent experiments. **p < 0.005 (unpaired two-tailed student t test).
Discussion
The present study demonstrated that IL-13Rα1 expression serves as a marker for M2 macrophages and influences their function. macrophages, which represent a heterogeneous population of cells with a wide range of phenotypic plasticity, play a major role in the development of innate and specific immunity, as well as in the resolution of inflammatory processes, among many other attributes [1]. Due to their functional heterogeneity and diverse anatomic localization, macrophages have been classified into subsets and termed with a variety of nomenclature [2, 38, 39]. Lately, macrophages were classified into 2 major subsets, with the M1 population as inflammatory and the M2 subset as anti-inflammatory [1, 2]. Generally, these designations were based on the expression of cell surface markers, the factors required for their differentiation, and/or the function of cytokines produced by the cells [1, 2, 40]. Herein, we demonstrate that IL-13Rα1 expression clearly distinguishes M2 (IL-13Rα1+) from M1 (IL-13Rα1−) macrophages. In addition, we show that the IL-13Rα1 chain associates with IL-4Rα to form a heteroreceptor that plays an important role in the differentiation, as well as in the function of M2 macrophages. Indeed, IL-13Rα1+ macrophages express surface markers that are usually associated with the M2 phenotype, including IL-4Rα, MR, FcγR and FcεRII [1, 2, 40], while IL-13Rα1− macrophages consistently display significant expression of IL-12Rβ2, which serves as a marker for M1 macrophages [6, 7, 12]. Aside from surface marker expression, IL-13Rα1+ macrophages express a functional IL-13Rα1/IL-4Rα heteroreceptor through which both IL-4 and IL-13 ligands stimulate up-regulation of genes characteristic of M2 cells, including Arg1 and DC-SIGN. This corroborates well with previous reports, which show Arg1 mRNA expression in macrophages is induced by IL-4 and IL-13 [41], which may be explained by expression of the heteroreceptor. IL-13Rα1− macrophages, however, lack expression of a functional heteroreceptor, are not able to utilize IL-13 for signaling, and thus fail to up-regulate either gene. In contrast, when IL-13Rα1− cells are stimulated with LPS, they up-regulate iNOS transcription, a characteristic associated with M1 macrophages [33, 42].
Further investigation revealed that when IL-13Rα1+ macrophages are stimulated with LPS, they produce IL-10, a functional hallmark of M2 macrophages, while IL-13Rα1− cells produce IL-6, IL-12, and TNFα inflammatory cytokines, which are functional attributes of M1 macrophages. IL-13Rα1+ macrophages are capable of inducing naïve CD4+ T cell differentiation towards the Th2 phenotype and exhibit enhanced phagocytic activity relative to IL-13Rα1− macrophages, which is characteristic of M2 macrophages [5].
Besides serving as a marker for M2 macrophages, IL-13Rα1 and the resulting heteroreceptor play a major role in the differentiation of macrophages from stem cells. Indeed, BM from IL-13Rα1−/− mice quantitatively yield more macrophages than stem cells from IL-13Rα1+/+ mice upon stimulation with GM-CSF. The majority of the resultant macrophages from IL-13Rα1− BM display an M1 phenotype, which suggests that in the absence of the heteroreceptor, M1 differentiation is more effective. Furthermore, stimulation with M-CSF yields a near equal number of macrophages in both strains and the frequency of M1 and M2 subsets was similar. Addition of cytokines to the differentiation cultures confirms signaling through the conventional IL-4R in IL-13Rα1-deficient macrophages, as evidenced by diminished iNOS gene expression with IL-4, but not IL-13 cytokine. Alternatively, both IL-4 and IL-13 seem to reduce differentiation into M1 macrophages when the BM cells originate from IL-13Rα1+/+ mice. The role of heteroreceptor expression in the differentiation and function of macrophages is further supported by the observation that IL-13Rα1−/− (M1) macrophages produce pro-inflammatory cytokines upon stimulation with LPS, while IL-13Rα1+/+ macrophages secrete the anti-inflammatory cytokine IL-10, which is in line with previous reports [43]. The role of the heteroreceptor in the function of M2 macrophages seems to be mediated through STAT6, based on the data that show similar degrees of STAT6 phosphorylation among IL-13Rα1+ and IL-13Rα1− with IL-4, both of which express the conventional IL-4 receptor (IL-4Rα/γc). However, when similar cultures are incubated with IL-13, the heteroreceptor expressed on IL-13Rα1+, but not IL-13Rα1− macrophages, triggers much higher STAT6 phosphorylation. The IL-13- heteroreceptor-mediated STAT6 phosphorylation produced spindle-shaped morphology in the M2 subset, a characteristic associated with M2 macrophages [44, 45]. IL-4 though, did not produce this morphology suggesting that its signaling through the heteroreceptor operates through a pathway distinct from IL-13 and thus has no effect on M2 Macrophage morphology.
Overall, this study identifies IL-13Rα1 as an additional cell surface marker of M2 macrophages and implicates the IL-13Rα1/IL-4Rα heteroreceptor in their differentiation and effector function. Given that M2 macrophages are involved in many clinical settings, targeting the heteroreceptor in these cells may prove useful against allergic and inflammatory diseases [46–50].
Materials and methods
Mice
C57BL/6 OTII mice, expressing a transgenic TCR specific for ovalbumin 323–339 (OVA323-339), have been described previously [36]. IL-13Rα1-GFP C57BL/6 mice expressing GFP under the control of IL-13Rα1 promoter, and IL-13Rα1-deficient C57BL/6 mice in which IL-13Rα1 gene expression was disrupted by deletion of exon 7, 8, and 9 have been previously described [21]. C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice were bred and maintained in our animal care facility for the duration of the experiments. All experimental procedures were performed according to the guidelines of the University of Missouri animal care and use committee.
Antigens
OVA323–339 peptide (SQAVHAAHAEINEAGR) was purchased from EZBiolab (Westfield, IN).
Production of extracellular IL-13Rα1
IL-13Rα1 cDNA encoding the extracellular domain of IL-13Rα1 (aa 27–339) with an N-terminal His tag was liberated from pQE-30 plasmid [51] by digestion with SphI and XhoI to liberate cDNA. This fragment was ligated into pFastBac plasmid (Invitrogen, Carlsbad, CA) to generate pFastBac-IL-13Rα1-EC, which was then used to generate baculovirus and to express extracellular IL-13Rα1 in both Sf9 and HiFive cells. IL-13Rα1 was purified using Ni-NTA agarose beads from QIAGEN. The elution fraction was tested for IL-13Rα1 by a Western blot using anti-RGS- HIS antibody (Qiagen) or by Coomassie stain.
Generation of the CTLL-2-MIG-IL-13Rα1 cell line
IL-13Rα1 cDNA was amplified from mRNA extracted from intestinal epithelium of BALB/c mice and cloned into the retroviral vector pMSCV-IRES-GFP (pMIG) [52]. To generate intact virus, 293 cells were transfected with pMIG-IL-13Rα1, or pMIG alone, with a packaging plasmid that allows 293 cells to release virions into the supernatant. IL-2-dependent CTLL-2 cells (a gift from Dr. Paul Allen, Washington University School of Medicine, St. Louis, MO) were infected with virion-laden supernatant from the 293 cells and GFP+ (CTLL-2-IL-13Rα1+) cells were purified using a MoFlo XDP sorter (Beckman Coulter, Fullerton, CA). CTTL-2 cells expressing truncated IL-13Rα1[53] were used for hybrid screening.
Generation of anti-IL-13Rα1 mAb 1G3
IL-13Rα1−/− BALB/c mice [21] were immunized subcutaneously (s.c.) in the footpads and axilla with 50 µg extracellular IL-13Rα1 emulsified in 200 µL phosphate buffered saline (PBS)/complete Freund’s adjuvant (CFA) (v/v). Mice were given a booster injection three weeks later with 50 µg of IL-13Rα1 emulsified in 200 µl PBS/IFA (v/v). Three days prior to fusion, mice were given 10 µg of IL-13Rα1 in PBS, intravenously (i.v). Spleen cells were fused with SP2/0 non-secreting myeloma cells using standard protocols. The hybridomas were then selected in HA media and 7-10 days after fusion supernatants were harvested and screened against CTLL-2-IL-13Rα1+ cells. Positive wells were cloned by limiting dilution and one clone designated 1G3 (IgG1, k), was selected for the study.
Antibodies
The antibodies used in this study were purchased from BD Biosciences, eBioscience, or R&D systems: anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD3 (500A2), F4/80 (BM8), anti-FcγRII/III (cl93), anti-c-Kit (CD117; 2B8), anti-CD11b (M1/70), anti-CD19 (1D3), anti-CD16/32 (2.4G2), anti-CD11c (HL3), anti-IL-4Rα (mIL4R-M1), anti-FcεRII (B3B4), anti-Ly-6G (RB6-8C5), anti-Ly6-C (HK1.4), anti-CD206(CO68C2), SiglecF (E50-24440), anti-pSTAT6 (pY641), anti-CD121a (Type1/P80), anti-I-Ab (NIMR-4), anti-CD80 (16-10A1), anti-CD86 (PO3.1), ST2L anti-IL-33 (FAB10041A), anti-IL-12Rβ2 (FAB 1959P), B220 (RA3-6B2) and FcεR1α (MAR-1).
Staining of CTLL-2-IL-13Rα1+ cells with purified anti-IL-13Rα1 mAb, 1G3
CTLL-2-IL-13Rα1+ cells (3 × 106 cells per well) were incubated with FcR blocking reagent (Miltenyi biotech) and purified 1G3 mAb (2 µg/mL) was added and incubated on ice for 30 minutes. Subsequently, excess antibody was removed by washing cells with buffer and bound anti-IL-13Rα1 antibody was detected with goat anti-mouse IgG (Fab')2-PE (1 µg/well). Antibody binding to the cells was determined by flow cytometry on a CyAn ADP analyzer (Beckman Coulter, Fullerton, CA) using FlowJo software version 8.8.6 (Tree Star).
Detection of IL-13Rα1 expression on adult and neonatal immune cells
Cells (1 × 106 cells/100 µL) from various organs of adult and neonatal IL-13Rα1+/+, IL-13Rα1−/−, and IL-13Rα1+/+-GFP C57BL/6 mice were incubated with mouse IgG Fc-fragment (Jackson ImmunoResearch) for 30 minutes on ice. Subsequently the cells were washed twice in FACS washing buffer and stained with antibodies against B220, CD3, CD4, NK1.1, FcεR1α, c-kit, CD11c, and CD11b. Also staining with 7AAD was used to exclude dead cells. The cells were then incubated with biotinylated anti-IL-13Rα1 (1G3) mAb (2µg/mL) on ice for 30 minutes. Streptavidin-PE was used to detect antibody binding to IL-13Rα1. Expression of IL-13Rα1 on each cell type was determined by gating on B220+ for B cells, CD3+CD4+ for T cells, NK1.1+ for NK cells, c-Kit− FcεR1α+ for basophils, c-Kit+ FcεR1α+ for mast cells, CD11c+ for DCs, and CD11b+ for myeloid cells.
Analysis of IL-13Rα1 expression in different organs
Whole cell lysates were prepared from tissues isolated from adult and neonatal mice (IL-13Rα1−/− and IL-13Rα1+/+), including intestinal epithelium, lungs, and liver, which were run on a gradient (4–12%) polyacrylimade NuPAGE gel, transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA) and blotted with anti–IL-13Rα1 mAb (IG3) and anti-β-actin (Abcam, Cambridge, MA) primary Abs. β-actin levels were used as loading controls. Goat anti-mouse IgG-HRP (Santa Cruz Bio) was used as secondary Abs.
Analysis of cytokine induced STAT6 phosphorylation
Purified CD11b+F4/80+IL-13Rα1+ (M2) and CD11b+F4/80+IL-13Rα1− (M1) macrophages (5 × 104/well) were stimulated with rIL-4 (30 ng), rIL-13 (30 ng), or medium alone for 30 minutes at 37°C in the presence of phosphatase inhibitors. NP40 cell lysates were then prepared and run on a gradient (4–12%) polyacrylimade NuPAGE gel. After transfer onto PVDF membrane, blotting was carried out with rabbit anti-STAT6, rabbit anti-pSTAT6 (Y641, Cell Signaling) and mouse anti-β-actin (2F1-1, Biolegend) primary Abs. Anti-rabbit IgG-HRP (Cell Signaling) and goat anti-mouse IgG-HRP (Sc-2005, Santa Cruz Bio.) were used as secondary Abs.
Co-immunoprecipitation
Intestinal epithelial cells from adult IL-13Rα1−/− and IL-13Rα1+/+ mice were lysed and incubated with purified anti-IL-4Rα (1 µg) (mIL4R-M1, BD Pharmingen), anti-IL-13Rα1 (2 µg) (1G3), or anti-β actin (0.5 µg) (2F1-1, Biolegend) at 4°C overnight. Protein G-Sepharose (Invitrogen) was then added for 1 h. The samples were spun down and the pellets were resuspended in NP0007 buffer (Invitrogen), boiled at 95°C for 5 minutes, and run on a gradient (4–12%) polyacrylimade NuPAGE gel. After transfer onto PVDF membrane, blotting was performed with anti-IL-13Rα1 (1G3), anti-IL-4Rα (mIL4R-M1), or anti-β actin primary antibodies. Goat anti-mouse IgG-HRP (Sc-2005) and goat anti-rat IgG-HRP (Sc-2006) were used as secondary antibodies (Santa Cruz Bio).
Cell sorting
Splenocytes from IL-13Rα1+/+-GFP neonatal and adult mice were stained with antibodies to CD11b, CD11C, and F4/80. CD11b+F4/80+ GFP+ (IL-13Rα1+) and CD11b+F4/80+ GFP− (IL-13Rα1−) macrophages were sorted on a Beckman Coulter MoFlo (Brea, CA), and only those with a purity of 95% or higher were used.
Differentiation of bone marrow cells into macrophages
This was performed according to previously described procedures [37]. Briefly, bone marrow cells were harvested from the femur of adult mice, filtered to remove bone fragments, and the RBCs were lysed. The cells were then washed, resuspended in culture media and incubated with either GM-CSF (10 ng/mL) or M-CSF (20 ng/mL). Seven days later, adherent cells were purified by positive selection using CD11b-conjugated magnetic beads and the MACS separation system (Miltenyi Biotec, Germany).
Cytokine ELISA
IL-4, IL-6, IL-10, IL-12, IFN-γ and TNF-α were detected according to BD Pharmingen’s (San Jose, CA) standard protocol using anti-cytokine antibodies. The OD450 was read on a SpectraMax 190 counter (Molecular Devices, Sunnyvale, CA) and analyzed using SOFTmax PRO 3.1.1 software. Graded amounts of recombinant cytokine were included for construction of the standard curve. Cytokine concentration was extrapolated from the linear portion of the standard curve.
Phagocytosis Assay
In vitro. Texas Red-conjugated zymosan A (S. cerevisiae) bioparticles (Z2843, Life Technologies) were used for phagocytosis assay as previously described [54]. Briefly, zymosan bioparticles were incubated with purified zymosan-specific rabbit polyclonal IgG antibodies for 1 h at 37°C for complex formation. Meanwhile, sorted CD11b+F4/80+ IL-13Rα1+ and IL-13Rα1− macrophages (10 × 104/well) were cultured in 96-well plates for 1 h at 37°C for adherence. The opsonin-bound bioparticles (1 × 104/well ) were then transferred to the culture and the incubation continued for 3 hrs. The cells were then detached and surface attached bioparticles were quenched by trypan blue solution. Phagocytized bioparticles were analyzed by flow cytometry.
In vivo. Texas-Red zymosan bioparticles (1 × 104 bioparticles in PBS/mouse) were injected intravenously into IL-13Rα1+/+-GFP mice for complement receptor-mediated internalization. After 24 hrs, splenocytes were harvested, stained with anti-CD11b, anti-CD11c, and anti-F4/80 antibodies and CD11b+ F4/80+IL-13Rα1+ macrophages were analyzed for presence of Texas-Red bioparticles by flow cytometry in comparison with CD11b+ F4/80+IL-13Rα1− macrophages.
T cell priming in vitro
Splenocytes from OTII mice were depleted of CD11c+ and CD11b+ cells and OVAp-specific CD4+ T cells were purified by positive selection on anti-CD4 antibody coupled microbeads (Miltenyi). The CD4+ T cells (3 × 105 cell per well) were then stimulated with irradiated CD11b+F4/80+IL-13Rα1+ or CD11b+F4/80+ IL-13Rα1− (3 × 104 cell per well) macrophages in presence of 10 µM OVA or HA peptide for 72 hrs. IL-4 and IFN-γ in culture supernatant were measured by ELISA.
RT-qPCR
IL-13Rα1+/+ and IL-13Rα1−/− C57BL/6 mice were used for generation of bone marrow-derived macrophages as well as for purification of splenic macrophages. The cells were stimulated either with media (Nil), IL-4 (30 ng/mL), IL-13 (30 ng/mL) or LPS (100 ng/mL) for 3 hrs and RNA was extracted using TRIZOL (Invitrogen/Life Technology, Carlsbad, CA). Quantitative RT-PCR was performed using the QantiTect Reverse Transcription kit from Qiagen (Valencia, CA) and StepOne Instrument Cycler (Applied Biosystems). The primers used were: iNOS: Forward 5’-CCAAGCCCTCACCTACTTCC-3’, Reverse 5’-CTCTGAGGGCTGACACAAGG-3, DCSIGN: Forward 5’-GGGAATTCAGAGTGGGGTGACATGAGTGAC-3’, Reverse 5’-CCCCAAGCTTGTGAAGTTCTGCTACGCAGGAG-3’, Arg 1: Forward 5’-CTCCAAGCCAAAGTCCTTAGAG-3’, Reverse 5’-AGGAGCTGTCATTAGGGACATC-3’, GADPH: Forward 5’-AACTTTGGCATTGTGGAAGG-3’, Reverse 5’-GGATGCAGGGATGATGTTCT-3.
Statistical Analysis
Data were analyzed using unpaired two-tailed Student t test using Prism Software v4.0c (Graphpad). Data were considered statistically significant when p < 0.05.
Supplementary Material
Acknowledgements
This work was supported by grants RO1NS057194, RO1AI048541, R21HD060089 (to H.Z.) from the National Institutes of Health. J.A.C. was supported by Life Sciences Fellowship from the University of Missouri, Columbia.
Abbreviations
- Arg1
arginase 1
- 7AAD
7-aminoactinomycin D
- DCSIGN
dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- FcεR
Fc epsilon receptor
- IL-4Rα
IL-4 receptor alpha
- IL-13Rα1
IL-13 receptor alpha1
- Macrophage
macrophage
Footnotes
Conflicts of interests: The authors declare no financial or commercial conflict of interest.
References
- 1.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends in immunology. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nature Reviews Immunology. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 3.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. Journal of leukocyte biology. 2006;80:1298–1307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nature immunology. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 6.Grohmann U, Belladonna ML, Vacca C, Bianchi R, Fallarino F, Orabona C, Fioretti MC, Puccetti P. Positive regulatory role of IL-12 in macrophages and modulation by IFN-γ. The Journal of Immunology. 2001;167:221–227. doi: 10.4049/jimmunol.167.1.221. [DOI] [PubMed] [Google Scholar]
- 7.Bastos KRB, Alvarez JM, Marinho CRF, Rizzo LV, Lima MRDI. macrophages from IL-12p40-deficient mice have a bias toward the M2 activation profile. Journal of leukocyte biology. 2002;71:271–278. [PubMed] [Google Scholar]
- 8.Stoermer KA, Burrack A, Oko L, Montgomery SA, Borst LB, Gill RG, Morrison TE. Genetic ablation of arginase 1 in macrophages and neutrophils enhances clearance of an arthritogenic alphavirus. J Immunol. 2012;189:4047–4059. doi: 10.4049/jimmunol.1201240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh Gh G. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J Leukoc Biol. 2002;71:597–602. [PubMed] [Google Scholar]
- 10.Dragomir AC, Sun R, Choi H, Laskin JD, Laskin DL. Role of galectin-3 in classical and alternative macrophage activation in the liver following acetaminophen intoxication. J Immunol. 2012;189:5934–5941. doi: 10.4049/jimmunol.1201851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ness TL, Ewing JL, Hogaboam CM, Kunkel SL. CCR4 is a key modulator of innate immune responses. J Immunol. 2006;177:7531–7539. doi: 10.4049/jimmunol.177.11.7531. [DOI] [PubMed] [Google Scholar]
- 12.Fantuzzi L, Puddu P, Varano B, Del Cornò M, Belardelli F, Gessani S. IFN-α and IL-18 exert opposite regulatory effects on the IL-12 receptor expression and IL-12-induced IFN-γ production in mouse macrophages: novel pathways in the regulation of the inflammatory response of macrophages. Journal of leukocyte biology. 2000;68:707–714. [PubMed] [Google Scholar]
- 13.Dasgupta P, Chapoval SP, Smith EP, Keegan AD. Transfer of in vivo primed transgenic T cells supports allergic lung inflammation and FIZZ1 and Ym1 production in an IL-4Ralpha and STAT6 dependent manner. BMC Immunol. 2011;12:60. doi: 10.1186/1471-2172-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrante CJ, Pinhal-Enfield G, Elson G, Cronstein BN, Hasko G, Outram S, Leibovich SJ. The Adenosine-Dependent Angiogenic Switch of macrophages to an M2-Like Phenotype is Independent of Interleukin-4 Receptor Alpha (IL-4Ralpha) Signaling. Inflammation. 2013 doi: 10.1007/s10753-013-9621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreider T, Anthony RM, Urban JF, Gause WC. Alternatively activated macrophages in helminth infections. Current opinion in immunology. 2007;19:448–453. doi: 10.1016/j.coi.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesce J, Kaviratne M, Ramalingam TR, Thompson RW, Urban JF, Jr, Cheever AW, Young DA, Collins M, Grusby MJ, Wynn TA. The IL-21 receptor augments Th2 effector function and alternative macrophage activation. J Clin Invest. 2006;116:2044–2055. doi: 10.1172/JCI27727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbert DR, Holscher C, Mohrs M, Arendse B, Schwegmann A, Radwanska M, Leeto M, et al. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity. 2004;20:623–635. doi: 10.1016/s1074-7613(04)00107-4. [DOI] [PubMed] [Google Scholar]
- 19.Sinha P, Clements VK, Ostrand-Rosenberg S. Interleukin-13–regulated M2 macrophages in combination with myeloid suppressor cells block immune surveillance against metastasis. Cancer research. 2005;65:11743–11751. doi: 10.1158/0008-5472.CAN-05-0045. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Nunez RT, L F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286:1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haymaker CL, Guloglu FB, Cascio JA, Hardaway JC, Dhakal M, Wan X, Hoeman CM, et al. Bone marrow-derived IL-13Ralpha1-positive thymic progenitors are restricted to the myeloid lineage. J Immunol. 2012;188:3208–3216. doi: 10.4049/jimmunol.1103316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Lee HH, Bell JJ, Gregg RK, Ellis JS, Gessner A, Zaghouani H. IL-4 utilizes an alternative receptor to drive apoptosis of Th1 cells and skews neonatal immunity toward Th2. Immunity. 2004;20:429–440. doi: 10.1016/s1074-7613(04)00072-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee HH, Hoeman CM, Hardaway JC, Guloglu FB, Ellis JS, Jain R, Divekar R, et al. Delayed maturation of an IL-12-producing dendritic cell subset explains the early Th2 bias in neonatal immunity. J Exp Med. 2008;205:2269–2280. doi: 10.1084/jem.20071371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graber PGD, Herren S, Aubry JP, Elson G, Poudrier J, Lecoanet-Henchoz S, Alouani S, et al. The distribution of IL-13 receptor alpha1 expression on B cells, T cells and monocytes and its regulation by IL-13 and IL-4. European Journal of Immunology. 1998;28:4286–4298. doi: 10.1002/(SICI)1521-4141(199812)28:12<4286::AID-IMMU4286>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 25.Hilton DJ, Zhang JG, Metcalf D, Alexander WS, Nicola NA, Willson TA. Cloning and characterization of a binding subunit of the interleukin 13 receptor that is also a component of the interleukin 4 receptor. Proceedings of the National Academy of Sciences. 1996;93:497–501. doi: 10.1073/pnas.93.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine & growth factor reviews. 2006;17:173–188. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Obiri NI, Debinski W, Leonard WJ, Puri RK. Receptor for interleukin 13. Interaction with interleukin 4 by a mechanism that does not involve the common gamma chain shared by receptors for interleukins 2, 4, 7, 9, and 15. J Biol Chem. 1995;270:8797–8804. doi: 10.1074/jbc.270.15.8797. [DOI] [PubMed] [Google Scholar]
- 28.Kawakami K, T J, Murata T, Puri RK. The interleukin-13 receptor alpha2 chain: an essential component for binding and internalization but not for interleukin-13-induced signal transduction through the STAT6 pathway. Blood. 2001;97:2673–2679. doi: 10.1182/blood.v97.9.2673. and Source, [DOI] [PubMed] [Google Scholar]
- 29.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annual review of immunology. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 30.Mueller TD, Zhang JL, Sebald W, Duschl A. Structure, binding, and antagonists in the IL-4/IL-13 receptor system. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2002;1592:237–250. doi: 10.1016/s0167-4889(02)00318-x. [DOI] [PubMed] [Google Scholar]
- 31.Kurowska-Stolarska M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, Pitman N, et al. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. The Journal of Immunology. 2009;183:6469–6477. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- 32.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 33.Ohmori Y, Hamilton TA. Requirement for STAT1 in LPS-induced gene expression in macrophages. Journal of leukocyte biology. 2001;69:598–604. [PubMed] [Google Scholar]
- 34.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 35.Leidi M, Gotti E, Bologna L, Miranda E, Rimoldi M, Sica A, Roncalli M, et al. M2 macrophages phagocytose rituximab-opsonized leukemic targets more efficiently than m1 cells in vitro. The Journal of Immunology. 2009;182:4415–4422. doi: 10.4049/jimmunol.0713732. [DOI] [PubMed] [Google Scholar]
- 36.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 37.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. The Journal of Immunology. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 38.Mebius RE, Kraal G. Structure and function of the spleen. Nature Reviews Immunology. 2005;5:606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 39.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 40.Mantovani A. From phagocyte diversity and activation to probiotics: back to Metchnikoff. European journal of immunology. 2008;38:3269–3273. doi: 10.1002/eji.200838918. [DOI] [PubMed] [Google Scholar]
- 41.Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, et al. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. The Journal of Immunology. 2001;167:6533–6544. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- 42.El-Gayar S, Thüring-Nahler H, Pfeilschifter J, Röllinghoff M, Bogdan C. Translational control of inducible nitric oxide synthase by IL-13 and arginine availability in inflammatory macrophages. The Journal of Immunology. 2003;171:4561–4568. doi: 10.4049/jimmunol.171.9.4561. [DOI] [PubMed] [Google Scholar]
- 43.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends in immunology. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto O, Hashiyama M, Minty A, Ando M, Suda T. Interleukin-13 selectively suppresses the growth of human macrophage progenitors at the late stage. Blood. 1995;85:3487–3493. [PubMed] [Google Scholar]
- 45.van der Plas MJ, van Dissel JT, Nibbering PH. Maggot secretions skew monocyte-macrophage differentiation away from a pro-inflammatory to a pro-angiogenic type. PLoS One. 2009;4:e8071. doi: 10.1371/journal.pone.0008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor alpha1 and the type II IL-4 receptor in asthma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:7240–7245. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramalingam TR, Pesce JT, Sheikh F, Cheever AW, Mentink-Kane MM, Wilson MS, Stevens S, et al. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. 2008;9:25–33. doi: 10.1038/ni1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cash E, Minty A, Ferrara P, Caput D, Fradelizi D, Rott O. Macrophage-inactivating IL-13 suppresses experimental autoimmune encephalomyelitis in rats. J Immunol. 1994;153:4258–4267. [PubMed] [Google Scholar]
- 49.Sinha S, Kaler LJ, Proctor TM, Teuscher C, Vandenbark AA, Offner H. IL-13-mediated gender difference in susceptibility to autoimmune encephalomyelitis. J Immunol. 2008;180:2679–2685. doi: 10.4049/jimmunol.180.4.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ochoa-Reparaz J, Rynda A, Ascon MA, Yang X, Kochetkova I, Riccardi C, Callis G, et al. IL-13 production by regulatory T cells protects against experimental autoimmune encephalomyelitis independently of autoantigen. J Immunol. 2008;181:954–968. doi: 10.4049/jimmunol.181.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnare M, Blum H, Juttner S, Rollinghoff M, Gessner A. Specific antagonism of type I IL-4 receptor with a mutated form of murine IL-4. J Immunol. 1998;161:3484–3492. [PubMed] [Google Scholar]
- 52.Yang L, Qin XF, Baltimore D, Van Parijs L. Generation of functional antigen-specific T cells in defined genetic backgrounds by retrovirus-mediated expression of TCR cDNAs in hematopoietic precursor cells. Proc Natl Acad Sci U S A. 2002;99:6204–6209. doi: 10.1073/pnas.092154599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orchansky PL, Ayres SD, Hilton DJ, Schrader JW. An interleukin (IL)-13 receptor lacking the cytoplasmic domain fails to transduce IL-13-induced signals and inhibits responses to IL-4. J Biol Chem. 1997;272:22940–22947. doi: 10.1074/jbc.272.36.22940. [DOI] [PubMed] [Google Scholar]
- 54.Kurotaki D, Kon S, Bae K, Ito K, Matsui Y, Nakayama Y, Kanayama M, et al. CSF-1–Dependent Red Pulp macrophages Regulate CD4 T Cell Responses. The Journal of Immunology. 2011;186:2229–2237. doi: 10.4049/jimmunol.1001345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.