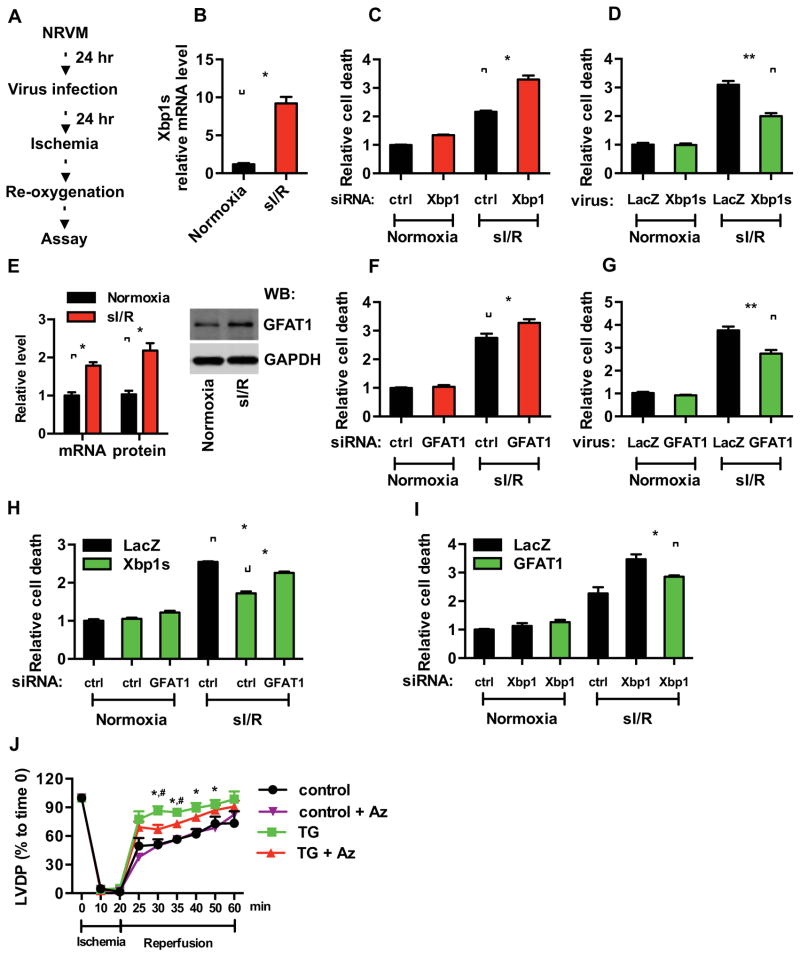
Figure 7. GFAT1 is required for Xbp1s-dependent cardioprotection during I/R.
(A) Experimental procedures for NRVM in vitro.
(B) Simulated I/R (sI/R) induced Xbp1s expression as assessed by qRT-PCR. N = 3.
(C) Knockdown of Xbp1 led to enhanced cell death in response to sI/R. Xbp1s expression was reduced by siRNA. Cell death was measured by LDH release. N = 3 for each group.
(D) Xbp1s overexpression protected cardiomyocytes from sI/R injury. NRVM were infected with lentivirus expressing either LacZ or Xbp1s. Cell death from sI/R was quantified by LDH measurements. N = 6.
(E) GFAT1 mRNA and protein levels were significantly induced in NRVM after sI/R. N = 3.
(F) Knockdown of GFAT1 exacerbated sI/R injury. GFAT1 expression was reduced by siRNA. NRVM were then subjected to sI/R, and cell death was assessed by LDH release. N = 3.
(G) GFAT1 overexpression protected cardiomyocytes from sI/R injury. NRVM were infected with lentivirus expressing either LacZ or GFAT1. Cell death from sI/R was quantified by LDH measurements. N = 6 for each group.
(H) GFAT1 knockdown diminished Xbp1s cardioprotection in sI/R. NRVM were first infected with LacZ or Xbp1s lentivirus. GFAT1 was reduced by siRNA. After sI/R, LDH assays were conducted. N = 3.
(I) Overexpression of GFAT1 significantly rescued cell death by Xbp1 silencing. NRVM were first infected with LacZ or GFAT1 lentivirus. Xbp1 was reduced by siRNA. After sI/R, cell death was quantified by LDH assay. N = 6 for each group. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01.
(J) Inhibition of GFAT1 diminished cardioprotection by Xbp1s. Control and TG mouse hearts were processed for Langendorff analysis (ischemia 20 min; reperfusion 40 min). Cardiac function was assessed as left ventricular developed pressure (LVDP). N = 7 for control, n = 4 for TG, n = 4 for control + Azaserine (Az), and n = 4 for TG + Az. Data are represented as mean ± SEM. *, TG vs control, p < 0.05. #, TG vs TG + Az, p < 0.05.
See also Figure S7.