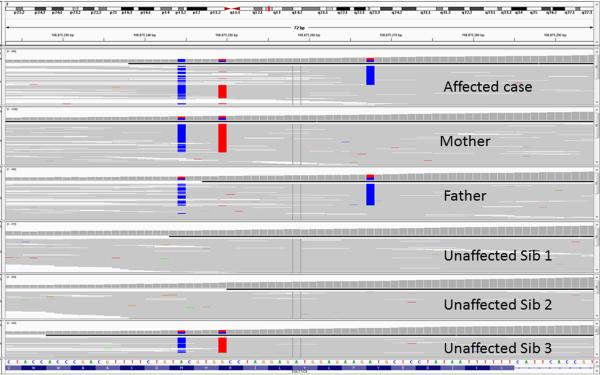
Figure 2. Integrated Genome Viewer Screenshot.
The Integrated Genome Viewer (IGV, http://www.broadinstitute.org/igv/) is a lightweight yet powerful tool for viewing short read pile ups. The example show includes pileups from six individuals: two parents, one affected child and three unaffected children. For convenience, a case was selected that shows two variants that are physically close to one another (and fit on the same screen). At the top of the display is a diagram of the chromosome being reviewed, with a small vertical red bar (between q12.1 and q13) highlighting the region being displayed below. The bulk of the display is taken up by six rows of pile-up data. Each row is an individual; each short read is a thin, gray horizontal line. Base positions that have been genotyped as non-reference are highlighted blue or red. In this case, the mother is heterozygous for two DNA variants. The father is heterozygous for one of the same variants and also for one different variant. The fact that each parent's pair of variants is cis-oriented is knowable because there are short reads with both variants, and short reads with neither variant. The affected sibling has DNA variations on both alleles, in contrast to any of the unaffected siblings.