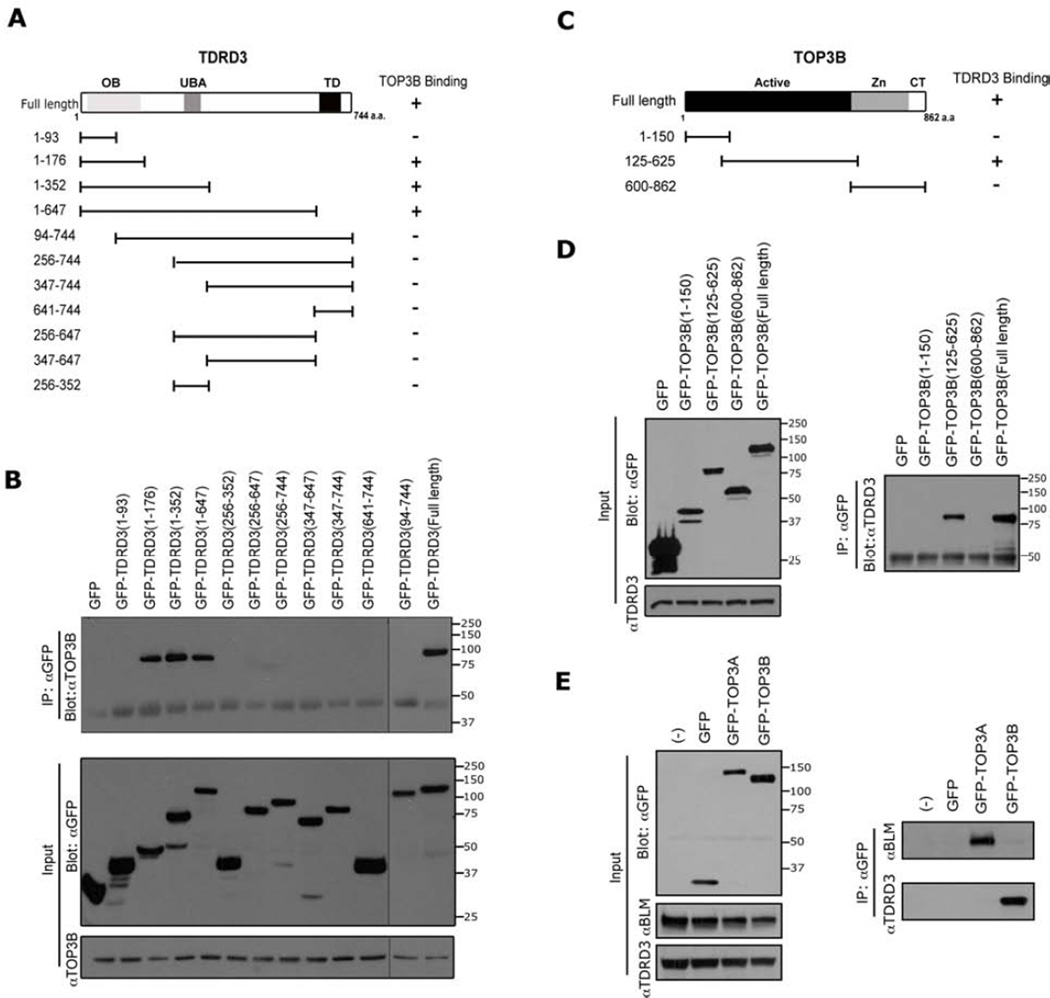
Figure 2. Mapping the interaction regions of TDRD3 with TOP3B.
(A) A series of GFP-fusion deletions of TDRD3 were generated. The locations of the OB-fold (OB), the ubiquitin-binding domain (UBA), and the Tudor domain (TD) are indicated. The graphic summary of the interactions observed in (B) is shown.
(B) A coIP assay was performed in HeLa cells transfected with the different TDRD3 GFP-fusion vectors. The cell lysates were immunoprecipitated with GFP antibody and eluted samples were blotted with αTOP3B. To observe all the samples, two membranes were spliced together. A gray line indicates the seam.
(C) Three GFP-fusion deletions of TOP3B constructs were generated. The locations of the catalytically active cleavage/strand passage domain (Active), the zinc-binding domain (Zn) and C-terminal domain (CT) are indicated. The graphic summary of the interactions observed in (D) is shown.
(D) A coIP assay was performed in HeLa cells transfected with the different TOP3B GFP-fusion vectors. Samples were prepared as described in (B) and blotted with αTDRD3 (right panel), and is shown (left panel).
(E) TDRD3 does not interact with TOP3A. HeLa cells were transiently transfected with GFP, GFP-TOP3A and GFP-TOP3B constructs. The cell lysates were immunoprecipitated with αGFP antibody and the eluted samples were assayed by Western blot analysis using αGFP antibody (left panel), and αTDRD3 and αBLM (right panel).