To The Editor:
Inhaled corticosteroids (ICS) are the primary anti-inflammatory therapy for the control and management of asthma, but their effects are characterised by some inter-individual variability that may have a genetic basis1–4. Identification of genetic markers that predict ICS treatment response will facilitate individualised treatment for asthma patients in the future, particularly in those with more severe disease. An association between genetic variation in GLCCI1 and response to ICS therapy in non-Hispanic white asthma subjects was observed by Tantisira et al.5 Genome wide association analysis of 118 trios (one asthmatic child and two parents) from the NHLBI Childhood Asthma Management Program (CAMP) identified 13 single nucleotide polymorphisms (SNPs) with evidence for association with the level of ICS treatment response. These 13 SNPs which included the GLCCI1 promoter polymorphism rs37972, were genotyped and evaluated in four additional independent collections: adult studies SOCS (Salmeterol Or CorticosteroidS) and SLIC (SalmeteroL ± Inhaled CorticosteroidS) (n=264); a second adult study (n=385); LOCCS (Leukotriene modifier Or Corticosteroid or Corticosteroid-Salmeterol) (n=185) and CARE (Childhood Asthma Research and Education) (n=101). In three of the four replicate populations GLCCI1 rs37973, a functional promoter polymorphism5 in complete linkage disequilibrium with rs37972 in white populations, was associated (P<0.05) with change in FEV1 (forced expiratory volume in one second) after 4–8 weeks of ICS treatment. The combined P value measuring association between rs37973 and ICS response over the four collections (n=935) was 0.0007. Tantisira et al. concluded that this functional GLCCI1 polymorphism, rs37973, was associated with response to ICS in asthma patients.
Using data from a recently completed genome wide association study of response to steroid therapy, we sought to confirm this observation in a homogeneous, well-characterised population of n=1,924 non-Hispanic white subjects by testing for association between rs37973 and measures of corticosteroid response in subjects treated with either fluticasone furoate (FF) or fluticasone propionate (FP) in seven GSK-sponsored clinical trials (NCT01165138; NCT01134042; NCT01086384; NCT01159912; NCT00603382; NCT00603278; NCT00603746). All seven studies were randomised, double blind, placebo controlled, parallel group multicentre studies in adolescent and adult subjects and employed change from baseline in FEV1 as their primary end-point, apart from HZA106837 which also investigated asthma exacerbations. HZA106837 (N=616) required each subject to have an asthma exacerbation within the 12 months prior to enrolment, whereas the other studies excluded subjects with previous asthma exacerbations. FEV1 was assessed at week 8 for all studies except HZA106837, which used week 12 data. Overall, except for FF and FP dose, baseline demographics and study characteristics were similar across all seven studies (Table 1).
TABLE 1.
Baseline, demographic and steroid response characteristics in seven GSK-sponsored clinical studies
| Clinical Studies | |||||||
|---|---|---|---|---|---|---|---|
| FFA109684 | FFA109685 | FFA109687 | FFA112059 | HZA106827 | HZA106829 | HZA106837 | |
| Clinical Phase | PIIb | PIIb | PIIb | PIIIa | PIIIa | PIIIa | PIIIa |
| N | 255 | 225 | 259 | 143 | 142 | 284 | 616 |
| Age (years) | 48.5 ± 13 | 42.1 ± 17.3 | 41.3 ± 15.9 | 42.4 ± 16.1 | 42.4 ± 16.0 | 47.3 ± 13.7 | 43.9 ± 16.5 |
| Age of Onset (Years) | 29.9 ± 17.9 | 25.0 ± 18.8 | 25.7 ± 18.5 | 25.3 ± 16.9 | 30.6 ± 18.5 | 33.6 ± 17.5 | 29.6 ± 18.1 |
| FEV1 at baseline (L) | 2.3 ± 0.6 | 2.4 ± 0.6 | 2.4 ± 0.7 | 2.4 ± 0.7 | 2.3 ± 0.6 | 2.2 ± 0.7 | 2.3 ± 0.7 |
| FEV1 at baseline (% of predicted) | 68.8 ± 11.5 | 73.7 ± 11.1 | 70.9 ± 12.2 | 72.5 ± 11.9 | 69.1 ± 10.4 | 66.3 ± 11.8 | 71.9 ± 10.5 |
| Run-In period | 28 day | 28 day | 28 day | 4 weeks | 4 weeks | 4 weeks | 2 weeks |
| Treatment on Run-In | ICS | ICS | non-CS | ICS | Asthma controller | ICS | FP or ICS |
| Sex (% female) | 56.5 | 59.6 | 58.7 | 55.9 | 63.4 | 59.5 | 66.2 |
| Height (cm) | 168.7 ± 9.5 | 168.3 ± 9.6 | 168.6 ± 10.1 | 168.8 ± 10.4 | 167.6 ± 9.0 | 167.9 ± 9.9 | 166.2 ± 9.7 |
| FF subject numbers | 199 | 182 | 210 | 71 | 142 | 142 | 616 |
| FF dose(s) mcg QD | 200, 400, 600, 800 | 100, 200, 300, 400 | 25, 50, 100, 200 | 100 | 100 | 200 | 100 |
| FP subject numbers | 56 | 43 | 49 | 72 | NA | 142 | NA |
| FP dose(s) mcg BD | 500 | 250 | 100 | 250 | NA | 500 | NA |
| Change FEV1 * | 0.18 ± 0.35 | 0.20 ± 0.42 | 0.29 ± 0.40 | 0.15 ± 0.36 | 0.33 ± 0.46 | 0.20 ± 0.44 | 0.19 ± 0.39 |
| ANCOVA Parameter Estimate** | 0.000 ± 0.030 | −0.017 ± 0.036 | −0.018 ± 0.033 | −0.034 ± 0.043 | 0.008 ± 0.050 | −0.007 ± 0.035 | −0.031 ± 0.022 |
| ANCOVA Estimate P value | 1.00 | 0.64 | 0.57 | 0.43 | 0.87 | 0.85 | 0.16 |
| GG to GA/AA Odds Ratio† | 1.62 | 1.24 | 1.61 | 4.00 | 0.63 | 1.08 | 1.33 |
| 95% confidence interval | 0.62–4.42 | 0.50–3.10 | 0.62–4.41 | 1.22–15.7 | 0.20–1.87 | 0.51–2.29 | 0.73–2.48 |
| P value | 0.33 | 0.65 | 0.33 | 0.02 | 0.40 | 0.85 | 0.35 |
FEV1 change was calculated as the pre-dose FEV1 measurement taken during at the clinic visit while still on-treatment minus the pre-dose FEV1 measurement taken on the first day of treatment
Analysis of covariance estimate of the rs37973 per-allele effect for each copy of the “G” allele, ± the standard error of the estimate. Model included five clinical covariates in addition to rs37973 coded as 0, 1, or 2.
Ratio of the odds of being in the lowest quartile of response (defined as change in FEV1, adjusted for five clinical covariates) given genotype GG relative to the odds given genotype GA or AA.
Germline DNA was extracted from peripheral blood collected from all 1,924 subjects all of whom provided consent for genetic analysis. Genotyping used either the KBiosciences Competitive Allele Specific PCR SNP genotype System (KASPar) (Hoddesdon, Herts, UK) or the Illumina Omni1-Quad panel (Expression Analysis, Durham, NC, USA). Analysis was undertaken in 1,916 subjects, including four with imputed data from rs37972. Eight subjects had missing covariate data.
Genetic association between rs37973 and ICS response was evaluated, with ICS treatment response defined as change from baseline in trough FEV1 using the last observation carried forward (in any subject who did not complete the specific trial), to week 8 or week 12 of FF or FP treatment. Change in trough FEV1 was regressed against covariates identified in this asthma population: age, percent of predicted baseline FEV1, study, height, asthma duration and drug (FF versus FP). We also evaluated the influence of rs37973 on subject placement within the highest and lowest response quartiles. Subjects who fell into the lowest (n=479) and highest (n=479) response quartiles were identified. Logistic regression was used to fit a model with quartile of response as the dependent variable, and genotype and relevant covariates as the independent variables. The P value measuring heterogeneity among the seven studies was 0.98, allowing pooling of data at the subject level.
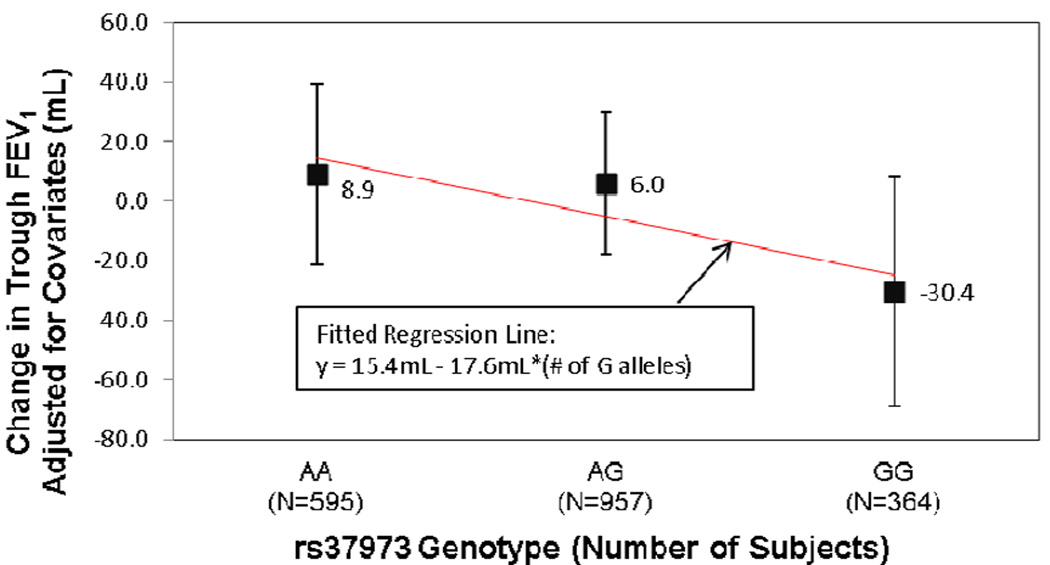
The minor allele frequency of rs37973 was 0.44 and its genotype frequencies were consistent with Hardy-Weinberg equilibrium (P=0.48). Covariate-adjusted FEV1 change was regressed on rs37973 genotype (Figure 1). Rs37973 did not influence change from baseline in FEV1 in this sample of 1,916 non-Hispanic white subjects treated with either FF or FP (P=0.15). However, this regression analysis suggested a trend toward a slightly lower ICS response for each additional copy of the rs37973 G allele. This direction of effect was consistent with that observed by Tantisira et al. In addition, the percentage change from baseline FEV1 (unadjusted for covariates, results not shown) was 10.4±0.8% in AA homozygotes and 8.8±1.0% in GG homozygotes, while the overall mean response was 9.8±0.4%.
FIGURE 1.
GLCCI1 rs37973 genotype does not significantly influence steroid response in 1,916 asthma patients
This genetic marker did not influence subject membership within response quartiles (P=0.08, Odds Ratio (OR) =1.39, 95% CI 0.96–2.00). The ORs in each clinical study ranged from 0.61 (HZA106827) (N=616) to 4.42 (FFA112059), which is one of two smallest clinical trials, N=143 (Table 1). Meta-analysis of the influence of rs37973 on subject placement within response quartile across all seven clinical studies, revealed similar results to the subject level pooled data, suggesting a non-significant trend towards lower ICS response in GLCCI1 rs37973 GG homozygotes, compared to AA homozygotes (P=0.11, OR 1.32, 95% CI 0.94–1.87). In order to closely mimic the analyses of Tantisira et al 5, change in trough FEV1 was also regressed against age, sex and height. All analyses in our sample were repeated using these covariates; there were no qualitative differences between the two sets of results.
Asthma is currently estimated to affect ~315 million people worldwide6. A robust genetic predictor of ICS response in asthma patients would provide clinical value7 as inter-individual variability in ICS treatment response is commonly observed. Tantisira et al 5 reported an association (P=0.0007) in a pooled analysis between GLCCI1 rs37973 and ICS treatment response as measured over 4–8 weeks in 935 white non-Hispanic adults and children, and an OR of 2.36 in a subject level pooled analysis evaluating subject placement with response quartiles. In this larger sample set, n=1,916, drawn from seven clinical studies, we did not confirm GLCCI1 rs37973 as a predictor of ICS response. However, the discrepant outcomes might have been due to various factors: the GSK studies were clinical trials specifically designed with FEV1 change as the primary endpoint for all studies except one, whereas those of Tantisira were designed around a range of other primary endpoints, including lung growth, time to treatment failure and the percentage of asthma control days; paediatric study participants were only included in the initial Tantisira5 cohort; and the duration of ICS treatment at the time of FEV1 assessment varied between the two evaluations. Further genetic studies will be required to fully elucidate the potential role of GLCCI1 in ICS treatment response in asthma patients.
Acknowledgment
EB and DM are funded by the following grants: NIH U01 HL65899; NIH RC2 HL101487.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflict of interest: LH, LF, SG, AY, LJ, and MM are employees of and hold stock in GlaxoSmithKline. ERB has served as a consultant for GSK and received consultant fees for other projects but not for this project. These clinical studies were funded by GlaxoSmithKline.
References
- 1.Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, Liggett SB, Gelfand EW, Rosenwasser LJ, Richter B, Israel E, Weschler M, Gabriel S, Altshuler D, Lander E, Drazen J, Weiss S. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Gen. 2004;13:1353–1359. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins GA, Lazarus R, Smith RS, Tantisira KG, Meyers DA, Peters SP, Weiss ST, Bleecker ER. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123:1376–1383. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pascual RM, Bleecker ER. Pharmacogenetics of asthma. Current Opin Pharmacol. 2010;10:226–235. doi: 10.1016/j.coph.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Howard T, Moore W, Ampleford E, Li H, Busse W, Calhoun W, Castro M, Chung K, Erzurum S, Fitzpatrick A, Gaston B, Israel E, Jarjour N, Teague G, Wenzel S, Peters S, Hawkins G, Bleecker E, Meyers D. Importance of hedgehog interactingprotien and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127:1457–1465. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua A, Himes BE, Lange C, Lazarus R, Sylvia J, Klanderman B, Duan QL, Qui W, Hirota T, Martinez F, Manger D, Sorkness C, Szefter S, Lazarus SC, Lemanske RF, Peters SP, Lima JJ, Nakamura Y, Tamari M, Weiss ST. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. New Engl J Med. 2011;365:1173–1183. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To T, Stanojevic S, Moores K, Gershon A, Batemen E, Cruz A, Boulet L-P. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health. 2012;12:204–211. doi: 10.1186/1471-2458-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maitland-van der Zee A, Raaijmakers J. Variation at GLCCI1 and FCER2: one step closer to personalised asthma treatment. Pharmacogenomics. 2012;13:243–245. doi: 10.2217/pgs.11.177. [DOI] [PubMed] [Google Scholar]