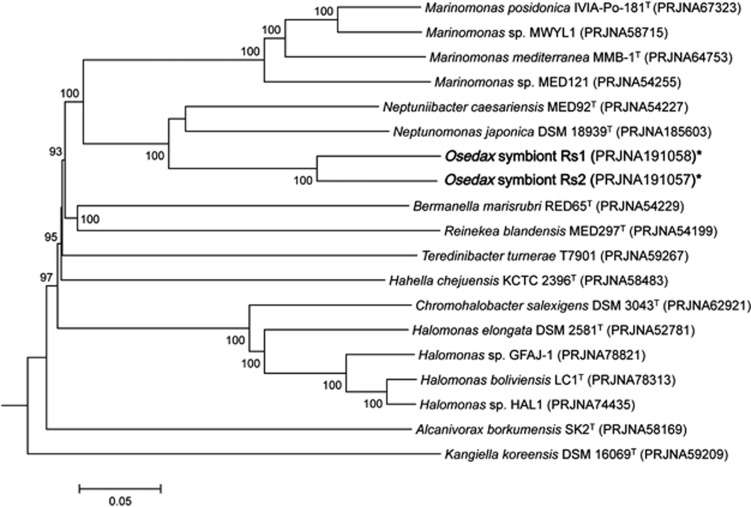
Figure 3.
Genomic relatedness of the Osedax symbionts to 17 other bacteria within the order Oceanospirillales available in public databases (an ‘*' denotes genomes from this study). The comparison is based on sequence similarity among a set of 300 highly conserved orthologous genes, showing greater than 40% amino-acid sequence similarity to Osedax symbiont Rs1. Sequences were aligned using CLUSTALX and multiple alignments were concatenated and then used to construct a genome tree using the neighbor-joining method implemented in MEGA. An evolutionary distance matrix for the neighbor-joining tree was generated according to the model of Jukes and Cantor (1969) and the resulting tree topology was evaluated by bootstrap analysis (numbers at nodes indicate bootstrap support from 1000 re-sampled data sets). The scale bar represents 0.05 substitutions per site.