Figure 3.
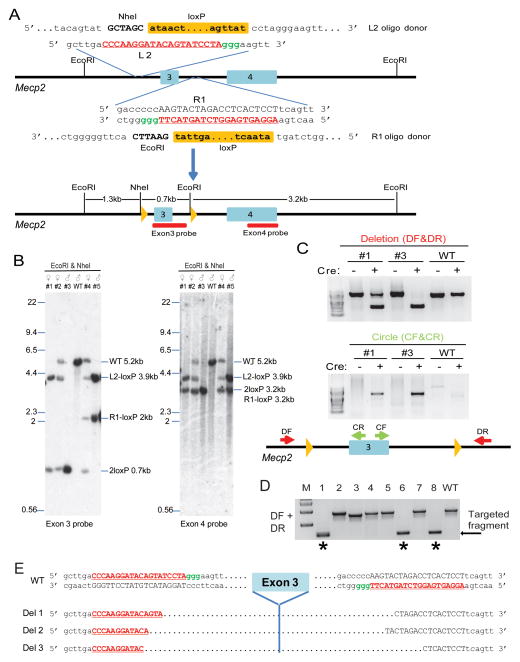
One step generation of a Mecp2 floxed allele. (A) Schematic of the Cas9/sgRNA/oligo targeting sites in Mecp2 intron 2 and intron 3. The sgRNA coding sequence is underlined, capitalized, and labeled in red. The PAM sequence is labeled in green. In the oligo donor sequence, the loxP site is indicated as an orange box, and the restriction site sequences are in bold and capitalized. The oligo contained 60bp homologies on both sides flanking the DSB. Restriction enzymes used for RFLP and Southern blot analysis are shown, and the Southern blot probes are shown as red boxes. (B) Southern analysis of targeted alleles. Data of five mice are shown. EcoRI/NheI-digested genomic DNA was hybridized with the exon3 probe. Expected fragment size: WT = 5.2 kb, 2loxP = 0.7 kb, L2-loxP = 3.9kb, R1-loxP = 2kb. The blot was then stripped and hybridized with the exon4 probe. Expected fragment size: WT = 5.2 kb, 2loxP = 3.2 kb. L2-loxP = 3.9kb, R1-loxP = 3.2kb. The sequence of the floxed allele is shown in Fig. S4b. (C) In vitro Cre-mediated recombination of the floxed Mecp2 allele. The genomic DNA of targeted mice #1 and #3 was incubated with Cre recombinase, and used as PCR template. Primers DF and DR flanking the floxed allele produce shorter products upon Cre-dependent excision. Primers CF and CR detect the circular molecule, which only form upon Cre-loxP recombination. The position of each primer is shown at the bottom cartoon. The deletion and circular PCR products were sequenced and the sequences are shown in Fig. S4c. (D) Injection of Cas9 mRNA and both L2 and R1 sgRNA generated Mecp2 mutant allele with deletion of exon 3. PCR genotyping using primers DF and DR identified defined deletion events in mice #1, #6, and #8 (indicated by stars). (E) Sequences of three mutant alleles with exon 3 deletions in three mice. R2 and L1 sgRNA coding sequences were underlined, capitalized, and labeled in red. The PAM sequence is labeled in green. See also Figures S3 and S4.