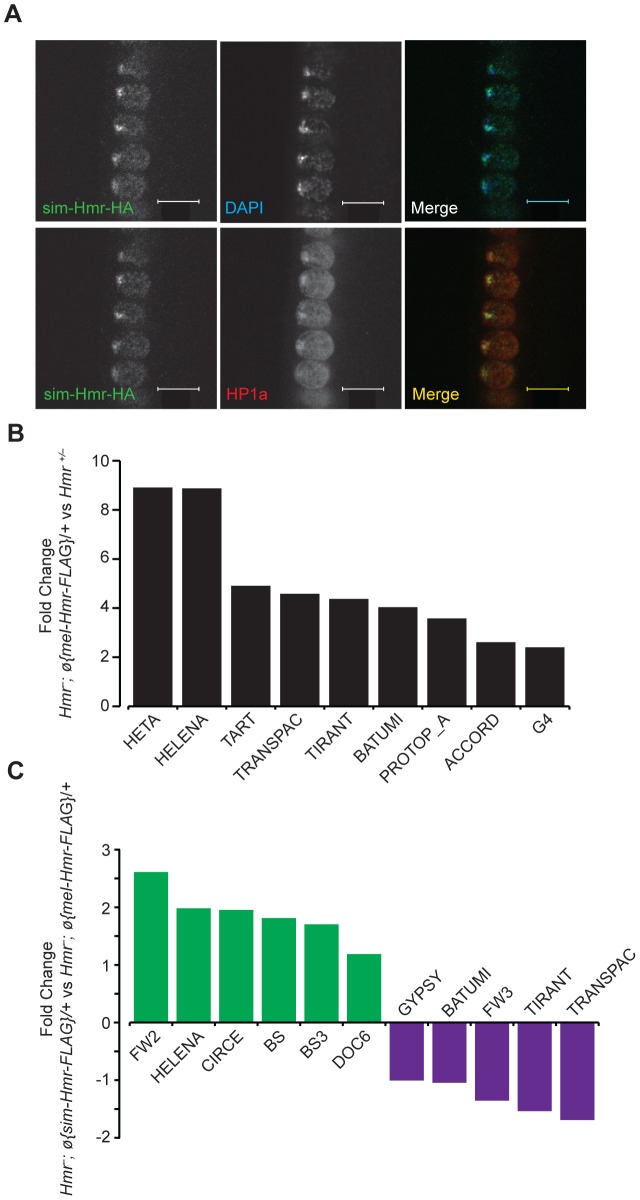
Figure 9. Hmr orthologs have diverged in their effects on a small subset of TEs.
(A) sim-Hmr-HA colocalizes with HP1a (red) in nuclear cycle 14 D. melanogaster Hmr3; sim-Hmr-HA embryos. The sim-Hmr-HA transgene was transformed into D. melanogaster at the identical attP2 site used for mel-Hmr-HA above (Figure 1). DAPI is shown in blue. (B) mel-Hmr-FLAG does not fully complement TE derepression in Hmr−. 9 TE families are 2–9× more highly expressed in Hmr−; ø{mel-Hmr-FLAG}/+ compared to Hmr+/−. (C) Comparison of TE expression in Hmr−; ø{mel-Hmr-FLAG}/+ and Hmr−; ø{sim-Hmr-FLAG}/+. For B and C, reads were mapped to the individual-insertion database. TEs are considered differentially expressed in the pairwise comparisons if there was at least a 2× fold change and p<0.001.