FIGURE 2.
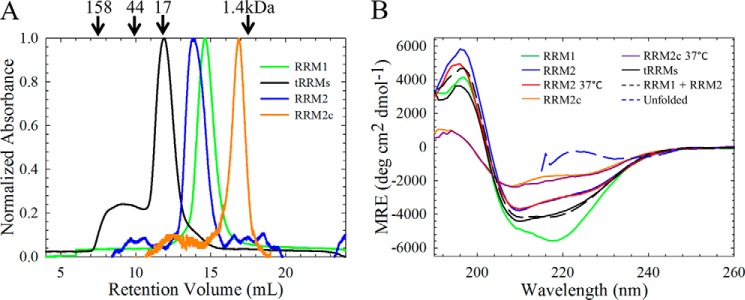
The isolated RRM domains are monomeric and contain significant secondary structure under physiological salt conditions. A, analytical size exclusion chromatography of the isolated (green, RRM1; blue, RRM2; orange, RRM2c) RRM domains reveals that each isolated domain is predominantly monomeric. The tethered domains (tRRMs, black) are also predominantly monomeric; however, some higher order species are also present, probably due to domain swapping. The arrows indicate retention times of molecular weight standards. B, RRM1 contains significant secondary structure compared with RRM2. RRM1 (green) has a minimum at 218 nm with a shoulder at 208 nm, whereas RRM2 (blue) has reduced ellipticity with a minimum at 210 nm and a shoulder at 220 nm. The cleavage fragment, RRM2c (orange) has further reduced ellipticity. All CD spectra were taken at 20 °C unless otherwise noted; physiological temperature (37 °C) does not alter the secondary structures of RRM2 (red) or RRM2c (purple). Tethering the RRMs (tRRMs; black) results in a CD spectrum resembling the sum of the isolated domains (black dashed line). The CD spectrum for each protein, reported as mean residue ellipticity as a function of wavelength, did not vary in the range of 5–60 μm, and all proteins showed similar unfolded spectra when denatured in 7 m GdnHCl (blue dashed line).
