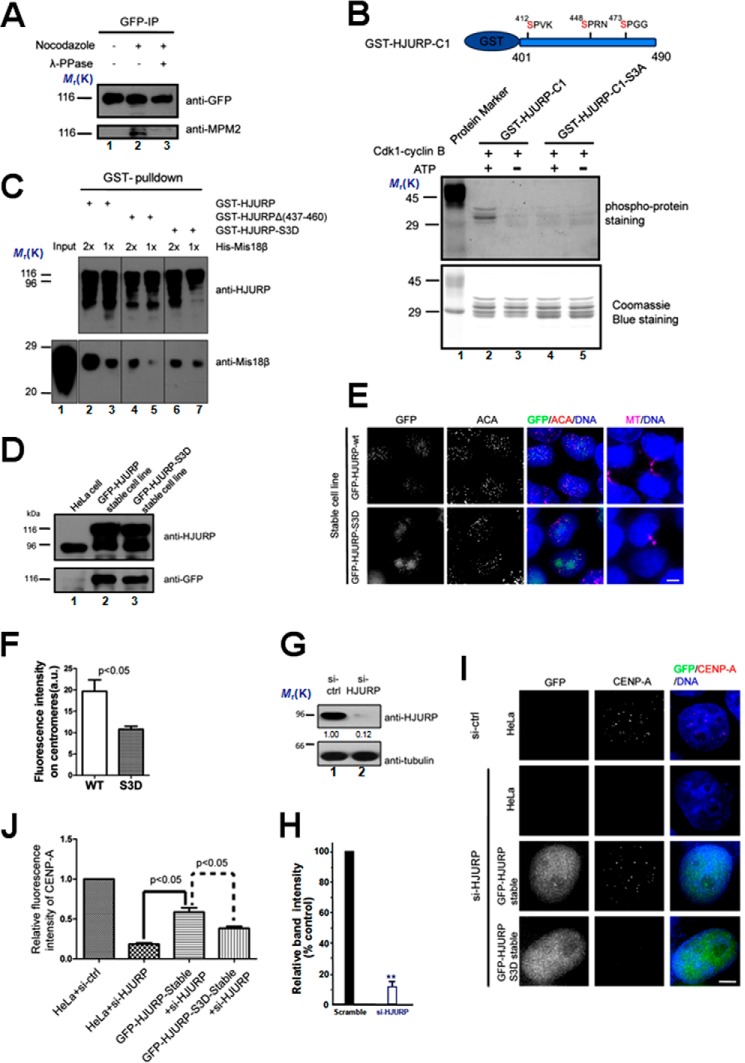
FIGURE 5.
HJURP recruitment to centromeres is regulated by CDK1. A, immunoprecipitation was performed using GFP antibody-coupled Sepharose beads using lysates from HeLa cells stably expressing GFP-HJURP treated as indicated. Nocodazole was added to block cells in mitosis. Extract of mitotic cells were treated with λ-phosphatase (λ-PPase) before immunoprecipitation to dephosphorylate proteins. The immunopurified products were probed with anti-GFP antibody and MPM2 antibody, respectively. B, an in vitro kinase assay was performed using CDK1 as kinase and purified GST-tagged HJURP fragment or the fragment harboring point mutations as illustrated above. The top panel shows the gel staining with a Pre-Q diamond phosphor-protein staining kit. The bottom panel shows Coomassie Blue-stained gel. C, different purified GST fusion HJURP mutants were used as affinity matrix to pull down purified His-Mis18β. The pull-downs were examined by anti-HJURP and anti- Mis18β antibody, respectively. D, immunoblots of lysates from normal HeLa cells, HeLa cells stably expressing GFP-HJURP-WT, and HeLa cells stably expressing GFP-HJURP-3D probed with the indicated antibodies. E, representative images of cells stably expressing GFP-HJURP or GFP-HJURP-S3D. Cells were fixed and co-stained for ACA (red), microtubule (magenta), and DNA (blue). Cells with midbody were selected to show that they were in early G1 phase. Scale bar, 5 μm. F, bar graph showing quantification of GFP fluorescence intensity at centromeres for the immunofluorescence assay described in D. The data represent mean ± S.E. (error bars) of three independent experiments. More than 70 centromeres (from 10 cells) were quantified for each group. Student's t test was used to calculate the p value for comparison of HJURP-3D and HJURP-WT. G, immunoblots of lysates from cells transfected with siRNA against HJURP or a scramble sequence siRNA (a negative control) probed with the indicated antibodies. 48 h after siRNA transfection, cells were collected and assayed by Western blotting. H, quantification of HJURP protein suppression by siRNA. Values represent the means ± S.D. (error bar) from three independent experiments. I, representative images of different cells as indicated treated with a control siRNA (si-ctrl) or siRNA against HJURP twice. 96 h after transfection, cells were fixed and stained for CENP-A (red) and DNA (blue). Scale bar, 5 μm. J, bar graph showing quantification of CENP-A fluorescence intensity at centromeres for siRNA treatments described in I. The data represent mean ± S.E. of three independent experiments. More than 200 centromeres (from 20 cells) were quantified per condition. Student's t test was used to calculate p value for comparison between the indicated groups.