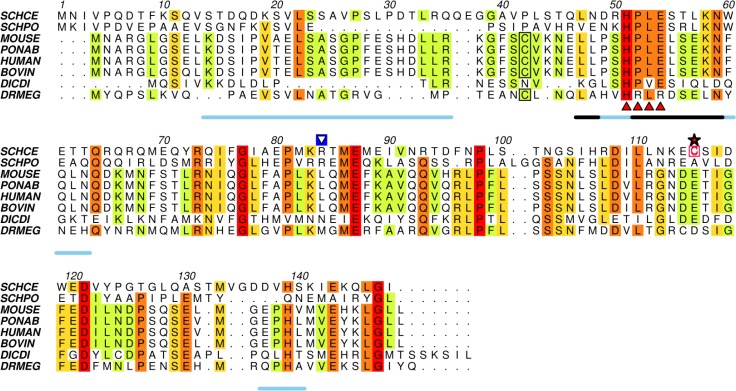
Figure 2.
The N-terminal half of Ump1 is predicted to be highly disordered. Amino acid sequence alignment of selected Ump1 proteins (see Table S1 for protein% similarities) was performed with ClustalW2, and rendered with Aline [35]. Disorder was predicted with RONN [30] for the selected amino acid sequences and a consensus line for disorder prediction (http://www.bioinformatics.nl/∼berndb/ronn.html) is printed below the alignment: the black line highlights residues where disorder is predicted for all the displayed sequences and the blue line represents regions where disorder is predicated for least 80% of the represented Ump1 orthologs. The position of the non-conserved Cys115 is indicated by a red star, the conserved motif HPLE is indicated by red triangles, and the Cys37 residue conserved in mammalian orthologs is boxed. The blue-boxed arrow above Arg84 points to one of the trypsin-cleavage sites identified by N-terminal sequencing after limited proteolysis experiments (Figure S1). SCHCE, Ump1 from Saccharomyces cerevisiae (UniProt accession code P38293); SCHPO, Ump1 from Schizosaccharomyces pombe (O74416); MOUSE, Ump1 from Mus musculus (Q9CQT5); PONAB, Ump1 from Pongo abelii (Q5R9L9); HUMAN, Ump1 from Homo sapiens (Q9Y244); BOVIN, Ump1 from Bos taurus (Q3SZV5); DICDI, Ump1 ortholog from Dictyostelium discoideum (Q55G18) and DRMEG, Ump1 from Drosophila melanogaster (Q9VIJ5).