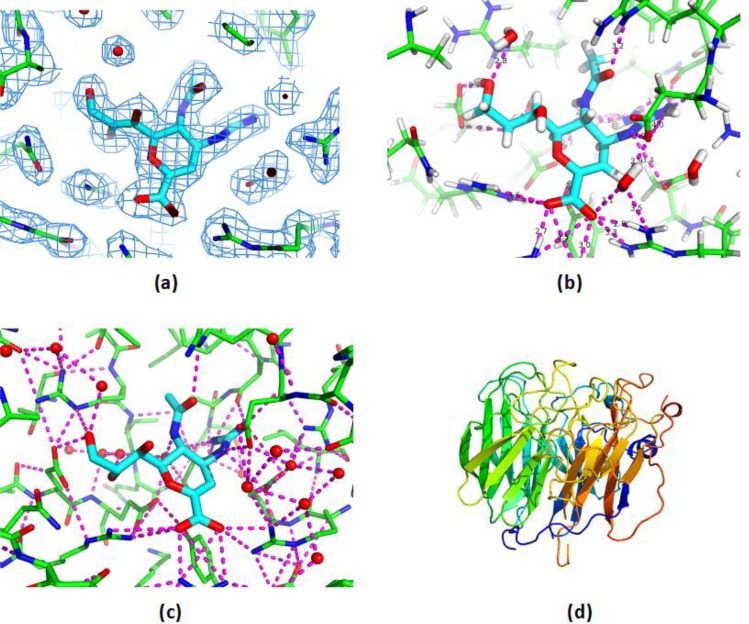
Figure 1.
Different representations of 3D protein structure provide different types of understanding. (a) The electron density map for ligand and surrounding binding site residues shows a good quality, rigid model. (b) Typical computational chemistry view, showing that protein-ligand binding involves many short hydrogen bonds. (c) A network view of all favourable polar interactions in the binding site region shows how the protein-ligand hydrogen bonds are parts of highly connected local environments, which also involve numerous bound water molecules. (d) Secondary structure view, suited for structural bioinformatics analysis. Protein is Neuraminidase in complex with Zanamivir. Images generated using PyMol (www.pymol.org), PDB Ids 2cml and 1nnc.