Abstract
Enzymes are an attractive alternative in the asymmetric syntheses of chiral building blocks. To meet the requirements of industrial biotechnology and to introduce new functionalities, the enzymes need to be optimized by protein engineering. This article specifically reviews rational approaches for enzyme engineering and de novo enzyme design involving structure-based approaches developed in recent years for improvement of the enzymes’ performance, broadened substrate range, and creation of novel functionalities to obtain products with high added value for industrial applications.
Keywords: protein engineering, rational design, de novo enzyme design, structure-guided engineering, promiscuous enzymes, artificial metalloenzymes
Introduction
The synthesis of chiral compounds is a challenge in chemistry. Enzymes are often superior to non-enzymatic catalysts concerning effectiveness, enantioselectivity and environmental friendliness. However, although natural enzymes are versatile biocatalysts catalyzing a wide range of chemical reactions, they are evolved towards the needs of their natural role. Thus, they are not available for many of the important conversions and substrates relevant for industry and do not fulfill the manifold requirements on enzymes used in industrial biotechnology. Enzymes should have high activity as well as high specificity and enantioselectivity towards frequently very challenging substrates. Moreover, they need to be stable during storage and resist a variety of - sometimes harsh - reaction conditions such as elevated temperature, extreme pH, high substrate / product concentrations and organic solvents. Thus, to fulfill all these criteria enzymes are nowadays routinely optimized by enzyme engineering for application in organic synthesis [1]. The published examples are uncountable and the interested readers are referred to numerous excellent reviews and book chapters available on protein engineering and the application of engineered enzymes in organic synthesis [2–9].
This article specifically reviews rational approaches for enzyme engineering and de novo enzyme design involving structure-based methods developed in recent years for improvement of the enzymes’ performance, broadened substrate range, and creation of novel functionalities to obtain products with high added value for industrial applications.
Current status of protein engineering
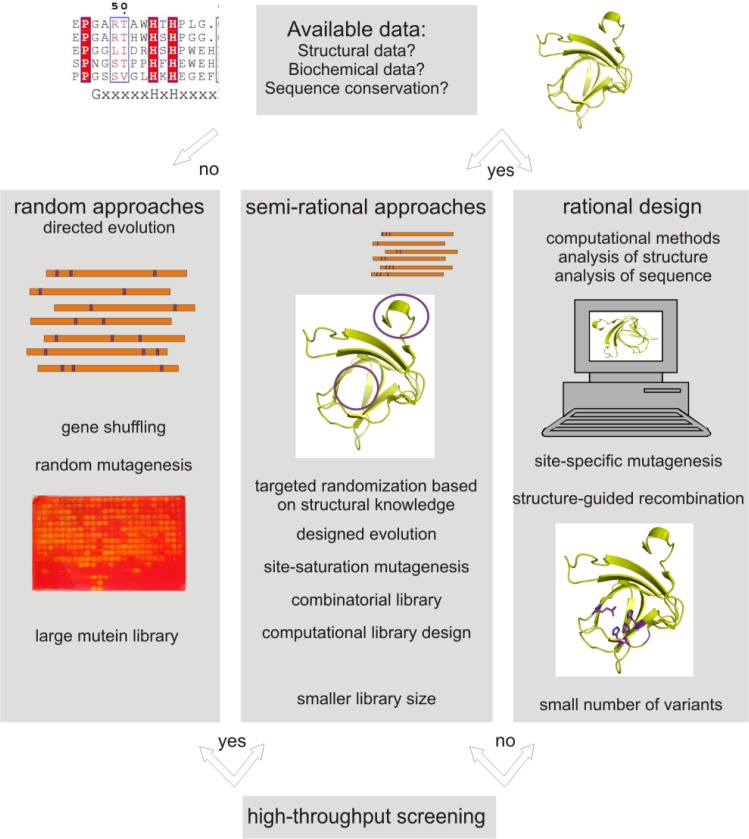
Generally there are two main strategies for protein engineering: directed evolution and rational design, which can be combined to semi-rational design or focused directed (designed) evolution (Figure 1).
Figure 1.
Overview of approaches for protein engineering by random, rational and combined methods.
Directed evolution can be achieved by two major approaches, either by randomly recombining a set of related sequences (e.g. gene shuffling), or by introducing random changes in single protein sequences (e.g. error-prone PCR). The advantage of directed evolution is that no structural information is needed and that variations at unexpected positions distant from the active site can be introduced. However, usually the changes are small and several rounds of evolution have to be applied and thus a high number of variants have to be screened, which is time and labor consuming and requires cheap, fast and reliable high-throughput assays.
With the availability of an increasing number of protein structures or reliable models, biochemical data and computational methods, enzyme engineering is developing more and more from random approaches (directed evolution) to semi-rational or rational (data-driven) design. In rational design biochemical data, protein structures and molecular modeling data are evaluated to propose mutations, which are introduced by site-specific mutagenesis. One of the advantages of a rational design approach is an increased probability of beneficial mutations and a significant reduction of the library size and thus less effort and time has to be applied for the screening of the library. This is especially advantageous if no high-throughput assay system is available.
Semi-rational design combines advantages of rational and random protein design creating smaller smarter libraries based on knowledge derived from biochemical and/or structural data [10]. An example for a semi-rational approach is CASTing (combinatorial active site saturation test), which uses the information derived from e.g. structural data to identify amino acids in interesting regions (e.g. active site), which are then mutated randomly or by site-saturation mutagenesis one by one or in combination [11–13, 8]. Random combination of mutations or correlated mutations at targeted positions can result in synergistic effects that might have been missed in single site-specific mutagenesis. However, these combinatorial approaches increase the library sizes tremendously and various computational methods have been developed in recent years, that help to decrease the library size by screening of virtual libraries and eliminating mutations predicted to be unfavorable for the protein fold [14–18].
Learning from natural diversity and conservation
Structure-guided consensus approach
In structure-guided consensus approaches sequence-based and structural data are combined. Generally, the consensus approach is based on the hypothesis that consensus amino acids of a sequence alignment contribute more than average to the fitness of the protein than the non-consensus amino acids. They proved to be favorable for the protein during natural evolution during which unfit variants are eliminated. Thus, changing non-consensus amino acids to consensus amino acids should improve e.g. thermostability (for details see [19]). A proof of concept was given after several rounds of this method, when the unfolding temperature of a fungal phytase was increased by more than 30°C to astonishing 90.4°C [20]. This approach was also successfully used to increase the thermostability of a cellulosomal endoglucanase, Cel8A, from Clostridium thermocellum 14-fold at 85°C without loss of catalytic activity [21]. However, after detailed analysis of the mutants, it turned out that of the eight identified consensus positions one single mutation (G283P) was sufficient to produce a thermostable variant. Glycine to proline mutations have been described before to increase the thermostability, however only at sites carefully selected [22, 23]. Thus, as not all of these consensus mutations actually contribute to stability and some might interfere with the enzymes’ activity, additional factors should be implemented. Structural features, such as the distance to the active site (improvements in stability in most cases require mutations further away from the active site, whereas changes of selectivity and activity usually target directly the active site), avoiding destabilization of helices or breaking existing hydrogen-bonds or salt-bridges, have to be considered [24–26].
By applying this structure-guided consensus approach, Bommarius and his group were able to increase the thermal stability of a penicillin G acylase (PGA) [25] and a glucose dehydrogenase (GDH) from Bacillus subtilis [27] by preserving the enzymes’ activities. In both cases about 50% of the variants showed increased thermostability. The three most stable mutations of GDH were successfully applied to two other GDHs from B. thuringiensis and B. lichenformis. Moreover, they showed that the most thermostable variants of GDH from B. subtilis were also more stable in high-salt solutions and homogeneous aqueous-organic media [28]. The same group combined the consensus approach with the B-fit (B-Factor Iterative Test) method to improve the thermostability of an α-amino ester hydrolase from Xanthomonas campestris resulting in a quadruple mutant (E143H/A275P/N186D/V622I) with 7°C improvement and 1.3-fold activity compared to wild-type [29]. In the B-fit method positions that display the highest flexibility (highest B-factor) in an enzyme's crystal structure are subjected to site-saturation mutagenesis [30–33]. The library size can be reduced by limiting the variation at the chosen positions to amino acids that are frequently present in an alignment using a consensus approach. An extension of the consensus approach is the combination of a sequence alignment with phylogenetic trees to identify the consensus sequence of a common ancestor [34]. It is thought that this common ancestor should be more stable due to the harsh environmental conditions during ancient times [35] and in addition this sequence might show promiscuous enzyme activity as it displays the turning point towards two separate enzymatic activities [36] (see below). Ancestral residues can also be introduced into modern sequences thereby combining the advantageous properties of the ancient (e.g. thermostability) and the modern proteins (e.g. activity, substrate range, selectivity) [37, 38].
3DM database
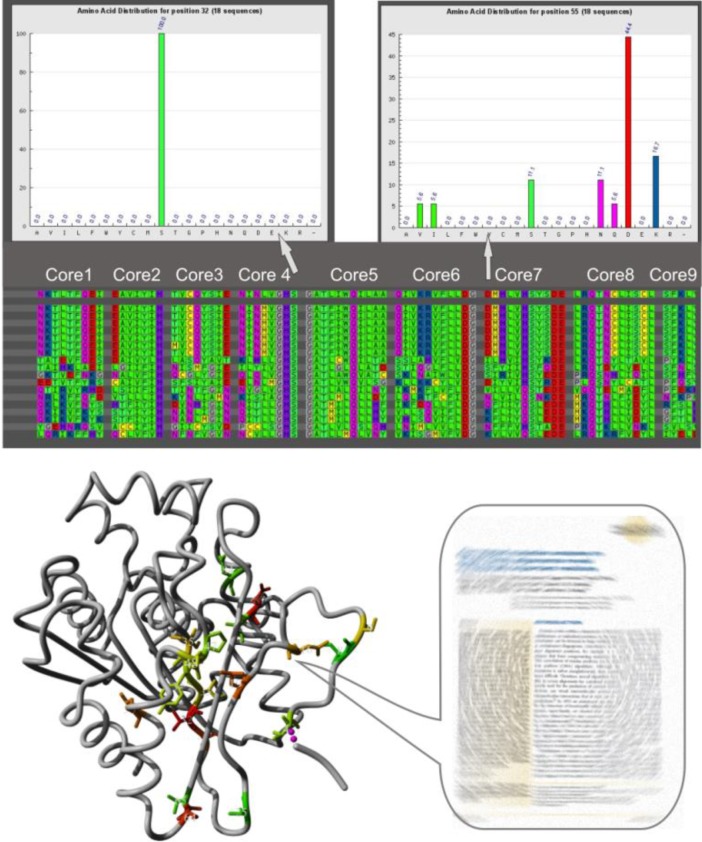
3DM databases are protein super-family platforms that combine different types of protein-related data including structures, multiple sequence alignments, conserved amino acids, (correlated) mutations, protein-ligand and protein-protein contact information. They are linked to literature containing mutations and biochemical data (Figure 2) [39–41]. 3DM databases can be applied for different purposes. Based on analysis of a 3DM database containing proteins of the α/β-hydrolase fold superfamily, the enantioselectivity (from E = 3.2 to E = 80) and activity (up to 240-fold) of an esterase from Pseudomonas fluorescens was improved by mutating four residues near the active site [42]. In addition, the thermostability of the same esterase was also improved in combination with the B-fit method [43]. Using the same database, a variant esterase from Paenibacillus barcinonensis with increased activity and enantioselectivity for the synthesis of tertiary alcohols was found [44]. Combination of the B-fit method with a 3DM database containing sequences of the glucosidase family 13 helped to identify positions where mutations led to increased thermal stability of a sucrose phosphorylase from Bifidobacterium adolescentis [45]. This study also revealed a synergistic effect of the two most flexible positions, as only pairwise mutations showed the desired outcome. In the most successful variant they introduced additional salt bridges. Analysis of a 3DM database for FAD-linked oxidases revealed, that almost all oxidases in the vanillyl-alcohol oxidase (VAO) fold subfamily contain a Gly or a Pro at a certain position, whereas other members that do not react with oxygen, have a different residue. Changing the corresponding residue in an L-galactono-γ-lactone dehydrogenase (GALDH) from Ala to Gly resulted in increased oxygen reactivity thus converting the dehydrogenase into an oxidase [46]. Very recently, a 3DM database for methyltransferases helped to identify amino acid residues relevant for catalytic function and binding of the cofactor S-adenosylmethionine, which were confirmed by mutagenesis analysis [47].
Figure 2.
Exemplified features of 3DM databases. 3DM databases link structural data, sequences and biochemical data (activity, stability, protein-ligand interaction, reported mutations). Middle: structure-sequence alignment of a subfamily, Top: amino acid distribution at chosen positions of the alignment, Bottom: mutations described in literature are linked to and depicted in a structure.
Structure-guided recombination
Another approach to improve the thermostability of proteins uses SCHEMA structure-guided recombination. SCHEMA is a computational algorithm, which estimates the disruption caused when amino acid residues that interact in the structure of a protein are recombined in chimeras. Sequences with low sequence similarity can be shuffled and the chimeras are rated according to the disruption caused [48–50]. With this method the thermostabilities of cellobiohydrolases class I, cellulases and cytochrome P450s were improved [51–53]. In addition, chimeric P450s were able to accept substrates that were not accepted by the parent protein, thus broadening the substrate range [54]. Recently, the thermostability of a Baeyer-Villiger monooxygenase (BVMO) phenylacetone monooxygenase was combined with the broader substrate range of other BVMOs by structure-guided subdomain exchange [55]. Interested readers can find more information about ‘Protein design with fragment databases’ in the recent review by Verschueren and coauthors [56].
Active site (re)design
Promiscuous catalytic enzyme activities
Generally, enzymes can be promiscuous concerning their reaction conditions (reaction condition promiscuity), their substrate range (substrate promiscuity), show an additional activity in the same active site (catalytic promiscuity) or due to a second active site (alternate site promiscuity) [57]. Some enzymes display a minor catalytic activity (accidental catalytic promiscuity) in addition to the main activity in the same active site. These low activities can be enhanced by mutagenesis and therefore provide a good starting point for protein engineering. On the other hand very similar enzymes, which belong to the same protein fold, often exhibit different enzyme activities. By comparison of the structure and the reaction mechanism of two enzymes, amino acids can be identified, the substitution of which could switch the catalytic activity of the enzyme (induced catalytic promiscuity). Some examples for increasing and introducing promiscuous catalytic activities are given below, for more extended reviews see [58–60, 57].
Based on the postulated reaction mechanism a single point-mutation was introduced into an arylmalonate decarboxylase from Alcaligenes bronchoseptimus to add a promiscuous racemase activity while retaining the decarboxylase activity [61]. Similarly, a detailed analysis of the reaction mechanism of the thiamine diphosphate dependent pyruvate decarboxylase identified a Glu473 residue, which was mutated and resulted in a 100-fold preference for the promiscuous carboligation reaction [62]. Hilvert and his group achieved the introduction of aldolase activity into a PLP-dependent racemase from Geobacillus stearothermophilus (thereby decreasing the racemase activity by >1000-fold) by mutation of just one amino acid (Y265A) in the active site, which removes a catalytic residue for the racemase activity, but creates space for the aldolase substrate [63, 64]. The aldolase activity was further extended by changing the same residue to a lysine [65]. Moreover, three additional mutations in the active site changed the preference to the (2R,3R) diastereomer relative to the (2R,3S) diastereomer. A double mutation in the active site abolished the racemase activity and increased the intrinsic forward half transaminase activity 6.6-fold in an alanine racemase from B. stearothermophilus [66].
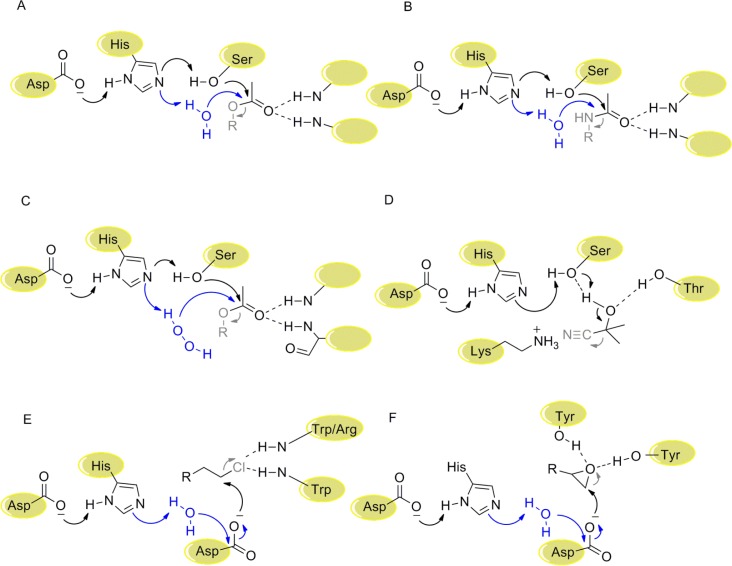
Enzymes with α/β-hydrolase-fold are one of the largest known protein families, including esterases, lipases, amidases, epoxide hydrolases (EH), dehalogenases and hydroxynitrile lyases (HNL) (Figure 3). Some of them already have intrinsic promiscuous enzyme activities [67].
Figure 3.
Reaction mechanism of A: esterase, B: amidase, C: perhydrolase, D: hydroxynitrile lyase, E: haloalkane dehalogenase, F: epoxide hydrolase. Esterase, amidase and perhydrolase share a common mechanism, in which a Ser, which is activated by a His-Asp dyad, serves as nucleophile. Subsequently, esterase and amidase are hydrolyzed by an activated water molecule, whereas in perhydrolases an activated H2O2 is involved. Haloalkane dehalogenase and epoxide hydrolase share a different mechanism, in which an Asp serves as nucleophile. A water molecule, which is also activated by the His-Asp dyad, hydrolyzes the ester bond between Asp and product. In contrast to these reaction mechanisms, in hydroxynitrile lyases no covalent enzyme-substrate intermediate is formed. Again the catalytic triad is involved, but the activated Ser acts as base and subtracts a proton from the hydroxyl group of the cyanohydrin. Subsequently the cyanide is released.
The first example in which a promiscuous activity seemed to be increased in an esterase is the perhydrolase activity of an esterase of P. fluorescens [68]. The perhydrolase activity of a single-mutant (L29P) apparently increased 28-fold from 0.24 to 6.8 U/mg, while the esterase activity decreased 100-fold from 14 to 0.14 U/mg. However, different substrates (acetic acid for the perhydrolysis and p-nitrophenylacetate for the hydrolase reaction) were used for the two reactions. Recently, by the use of the same ester substrate, methyl acetate, for both reactions, it was shown that mutation L29P actually decreased the perhydrolase activity 15-fold and the hydrolysis 3-fold. Moreover, experiments revealed that the mutant did not have higher selectivity for H2O2 than for water as it was originally proposed, but that in the mutant the acyl-enzyme intermediate with acetic acid is formed faster than in the wild-type [69].
Both, esterases and some HNLs, belong to the α/β-hydrolase superfamily and have the typical conserved Asp, His and Ser catalytic triad in their catalytic site. Although the substrates and the reaction mechanisms are different, two groups managed to introduce HNL activity into esterases. Schwab and his group converted the bacterial esterase EstC from Burkholderia gladioli into an HNL; one mutation (S276K) was sufficient to generate HNL activity and to abolish esterase activity [70]. Kazlauskas and his group switched the plant esterase SABP2 activity by just two point mutations (G12T and M239K), resulting in an enzyme with strongly reduced esterase activity, but clearly detectable HNL activity with racemic mandelonitrile (20 mU/mg, kcat/KM = 72 min-1M-1), but with low enantioselectivity cleaving (R)- and (S)-mandelonitrile with 12.6 and 15.5 mU/mg, respectively. In the synthesis direction, the (S)-product showed only 20% ee [71]. Comparison of the structures of epoxide hydrolases with esterases revealed differing loops at the entrance to the active site. Subsequent exchange of the loop of the esterase by an epoxide hydrolase loop finally introduced EH activity into an esterase from P. fluorescens (kcat 0.01 s-1, [72]). The best variant showed high enantioselectivity (E > 100) for the (R)-enantiomer of p-nitrostyrene oxide. Some esterases already display promiscuous amidase activity. However, so far no successful shift in the reaction preference has been reported [73]. Very recently, the promiscuous enantioselective (γ)-lactamase activity of (2-azabicyclo[2.2.1]hept-5-en-3-one) in the P. fluorescens esterase I was increased 200-fold by the introduction of a point mutation (L29P) and the structural and mechanistic determinants for the catalytic promiscuity and enantioselectivity were identified by molecular modeling [74]. Interestingly, this is the same mutation as has been reported to play a role in perhydrolysis reaction [68] (see above). These studies together with investigations of Syren and Hult on the prerequisites for amide hydrolysis [75] might lead to conversion of an esterase into an efficient amidase.
Lipase B from Candida antarctica (CAL-B) was analyzed by quantum mechanical simulations for the possibility to introduce aldolase activity. Site-directed mutagenesis of Ser105 in the active site proved that very low activity for an aldol reaction was achieved [76]. Interestingly, the S105A variant is also able to catalyze Michael-type additions faster than the wild-type [77, 78]. It was suggested that the mutation of the Ser residue, which is part of the catalytic triad in the natural hydrolase activity, disrupts the hydrolase activity with no effect on the Michael-type reaction with an ester substrate [79]. Recently, docking studies of methyl 2-methyl-2-nitro-5-oxohexanoate in CAL-B S105A revealed that theoretically there is no preferred binding mode for either the (R)- or the (S)-enantiomer in the voluminous active site explaining the lack of enantioselectivity [80].
Despite the increasing number of redesigned active sites to introduce or improve promiscuous activities, most of them still lack behind natural enzyme activities. Care has also to be taken, that the apparent promiscuous reaction de facto stems from enzymatic catalysis and is not chemically catalyzed by a prosthetic group only [81] or by amino acids not related to an active site (e.g. EstB from B. gladioli, [82]) or from an apparent new reaction due to an overlooked change of the actual substrate e.g. hydrolysis of substrate prior to the measured reaction [83]. Thus, the choice of an appropriate method for analysis and proper controls are crucial.
Broadening substrate range
Nowadays substrate docking is a routine method in enzyme engineering to carve the active site to improve enzyme activity, broaden the substrate scope towards industrially interesting compounds and increase or invert enantioselectivity. A few representative examples are given in this review.
In 2005, the (R)-selective hydroxynitrile lyase from Prunus amygdalus was one of the first lyases to be redesigned by modeling to give enzyme variants with improved enantioselectivity (>96%) for the synthesis of (R)-2-hydroxy-4-phenylbutyronitrile, which is an intermediate of angiotensin-converting enzyme inhibitors (ACEi) known as prils [84]. The crystal structure of another hydroxynitrile lyase from Manihot esculenta revealed that a bulky tryptophan at the entrance of the active site might be the reason for the preference of small substrates, which was confirmed by site-directed mutagenesis to a small Ala residue resulting in enhanced activity [85, 86]. Interestingly, mutation of this Trp residue to smaller residues (Ala, Met, Phe) was also identified in the highly similar HNL from Hevea brasiliensis by directed evolution (Schwab, unpublished results). Other examples concerning engineering of HNLs are given in other reviews (e.g. [87, 87b]). Similarly, molecular dynamics simulation and structure-based enzyme design identified key functional residues in the active site access tunnel of a dehalogenase from Rhodococcus rhodochrous [88]. Substitutions in these positions resulted in variants with 32-fold increase of activity towards 1,2,3-trichloropropane (TCP). This improvement was shown to originate from changes in solvent accessibility of the active site. Analysis of a transition-state model of 1-phenyl-1-hexanol in a lipase from Burkholderia cepacia proposed three amino acids that might improve the activity and enantioselectivity towards secondary alcohols with bulky substituents on both sites [89]. Experiments confirmed that indeed a double mutant of two of these residues showed high conversion and high enantioselectivity (E > 200, compared to E = 5 before). Analysis of the structure and molecular modeling allowed the identification of amino acids critical for switching the substrate specificity of a Drosophila melanogaster 2’-deoxynucleoside kinase from thymidine to 3’-deoxythymidine by side-directed mutagenesis [90]. In an approach combining in silico modeling, site-saturation mutagenesis and directed evolution, Savile and coworkers created an ω-(R)-transaminase, which is able to convert the bulky-bulky pro-sitagliptin ketone (200 g/L) to sitagliptin (an active pharmaceutical ingredient API) of >99.9% ee with 92% yield in 50% DMSO after eleven rounds of mutagenesis inserting a total of 27 mutations in the most active mutant [91]. The activity for aliphatic amines was improved by replacing a bulky tryptophan residue in the active site of an (S)-selective ω-transaminase from Vibrio fluvialis by a glycine to create a larger substrate binding pocket [92]. Using homology models Berglund and his group designed variants of originally (S)-selective ω-transaminases from Chromobacterium violaceum and Arthrobacter citreus that showed improved or reversed enantioselectivity [93, 94]. For other transaminase engineering approaches see Mathew and Yun [95]. By combination of directed and designed evolution (site-saturation mutagenesis of three amino acids in the active site, that have been identified in the structure) of the esterase EstB from B. gladioli the enantioselectivity was inverted [96]. Simultaneous site-saturation mutagenesis of two residues in the active site of esterase B2 from B. subtilis resulted in inverted enantioselectivity from ER > 100 to ES = 65 [97].
Many oxidoreductase enzymes are NAD(P)H dependent. In pathway engineering of microorganisms used for the production of industrially relevant products the balance of cofactor consumption and regeneration is essential to obtain a high yield. Structure-guided site-specific mutagenesis to change the cofactor preference has been reported several times e.g. in xylose reductases [98, 99], phosphite dehydrogenase [100], alanine dehydrogenase [101], (BVMO) phenylacetone monooxygenase [102] and many more. As nicotinamide cofactors are expensive, they are frequently recycled in biotechnological processes, e.g. by the use of NAD(P)H oxidases. Bommarius and his group engineered the substrate binding pocket of an NADH oxidase from Lactobacillus plantarum by site-specific mutagenesis to accept also NADPH [103].
De novo enzyme design
Novel enzymes can be either designed by recreating known enzymatic functions in proteins with a different fold or by introduction of activities that have not been observed in natural enzymes before into a chosen protein scaffold. The biggest challenge, however, is the design of de novo enzymes that are not based on natural sequences, thus designing the complete protein from scratch [105–108]. This is beyond the scope of this review and will not be addressed in detail. Most de novo enzyme design approaches depend on computational methods, which have been developed and improved in the last two decades [108, 109, 6, 106, 111–113].
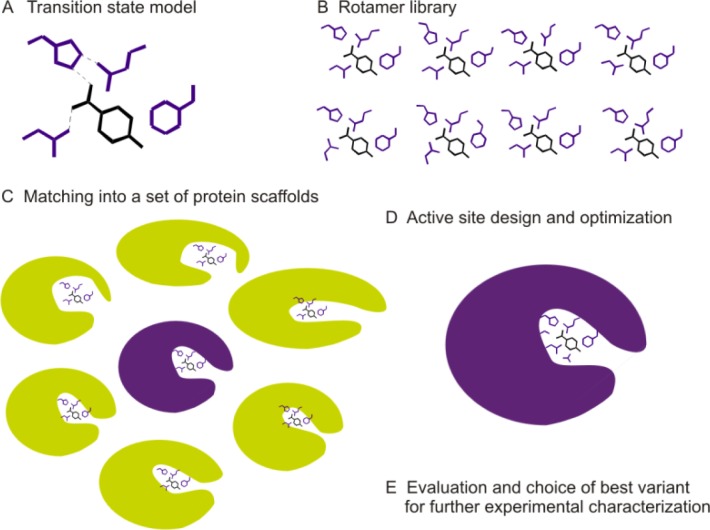
For the approaches described here, a detailed knowledge of the reaction mechanism and transition state is crucial to be able to predict which amino acids at which positions and distance are necessary to form an active site and catalyze the desired chemical reaction. In an approach by the Baker lab, in the first step idealized models of an enzymatic transition state are created by the use of quantum mechanical calculations (theozymes), which are in the next step fitted into protein scaffolds, which were identified from a library of folds and can accommodate the amino acids of the active site without clashes (RosettaMatch) [113, 114]. However, usually no perfect match is found and thus subtle deviations from the theozyme geometry have to be accepted (Figure 4). Another possibility offers SABER (Selection of Active/Binding sites for Enzyme Redesign), a computational method developed by the Houk lab, which searches for structures in which the necessary amino acids are already in place and only the substrate needs to be fitted in its transition state geometry [115]. Finally, in both approaches, the surrounding side chains are optimized for favorable interactions with the substrate/transition state model and stabilization of the protein fold (RosettaDesign) [113, 114].
Figure 4.
De novo design of active sites in existing protein scaffolds.
The most prominent examples for designed novel enzymes with non-natural enzyme activities are a Kemp eliminase, a Diels-alderase and a retroaldolase. Although currently enzymes, which are designed de novo by computational methods don't reach the performance of natural enzymes or variants evolved in the laboratory, in some cases, they can be improved by standard protein engineering methods [5, 116]. Mechanistic studies and structural analysis (also of inactive designs) will give indications about the quality of the design, identify problems, and help to improve the model and the computational parameters [117–124].
The computationally designed retroaldolase utilizes a catalytic lysine residue that forms a Schiff base with a keto group on the substrate, and then serves as an electron sink during C-C bond cleavage. To date 65 active designs were constructed in 14 different protein scaffolds [124, 125], however with very low starting activities (kcat/KM values of less than 1 M-1s-1), which were further improved by systematically mutating amino acids at or near the active site (improve of kcat/KM values up to 88-fold) and directed evolution (in one protein the activity improved 700-fold over ten evolutionary cycles, a combination of a 100-fold increase in kcat/KM and an increased yield of soluble protein) [124, 121]. However, the best improved and evolved retroaldolase design reaches a kcat/KM value of 55, which is still below catalytic antibodies with kcat/KM value of 490 under the same reaction conditions [124].
Other examples in which the initial design was further improved by random mutagenesis are the Kemp eliminase KE07 (improvement of kcat/KM values from 12 to 2,600 M-1s-1) [126] and the variant KE70, which was improved by a combination of rational and random mutagenesis to kcat/KM values up to 5x104 M-1s-1 [127]. The initial design combined a catalytic base, either Asp or Glu (KE07), or a His-Asp/Glu dyad (KE70), a hydrogen bond donor (Ser or Lys) to stabilize the accumulating negative charge that develops on the phenolic oxygen, and an aromatic residue for substrate binding and delocalization of the negative charge of the transition state. Of the 59 final designs, only eight showed activity [128]. Interestingly, the initially catalytically most active design KE59 was one of the most instable variants. After introduction of fold-stabilizing consensus mutations and 16 rounds of directed evolution the kcat/KM value increased > 2000-fold (from 163 to up to 0.6x106 M-1s-1) due to a large improvement of kcat [129]. Moreover, the optimized variant has a broader substrate acceptance. Structural analysis and MD simulations of improved variants of KE07, KE70 and KE59 led to explanations for the higher activity including a tighter substrate binding and the stabilization of the active-site dyad (KE70) in a conformation optimal for catalysis and a general more soluble and stable protein. In a different strategy of the Houk and Mayo labs - instead of characterizing many different designed proteins - the active site of a single template protein was redesigned to a Kemp eliminase using an approach described before [130]. After a detailed analysis of the structure and dynamics of an inactive first design and identification of possible causes for the inactivity, the focus of the design was moved from the native active site of the protein to a small pocket deeper in the protein [131]. This second design contained twelve mutations and resulted in a Kemp eliminase with a kcat/KM value of 123 M-1s-1, which was further improved 3-fold by single point mutations. Interestingly, two other groups managed to introduce Kemp eliminase activity into a hydrophobic pocket of calmodulin and a buried cavity in T4 lysozyme (differing from the native active site) by the introduction of just one charged amino acid, Glu and His respectively; however exhibing lower activity (kcat/KM values of 5.8 and 1.8 M-1s-1, respectively) than the above mentioned designs [132, 133]. The kcat/KM values of catalytic antibodies for Kemp eliminations were reported to be 5,500 M-1s-1 using carboxylate as general base [134] and what is even more intriguing serum albumins e.g. BSA can catalyze the same reaction with kcat/KM values of 2,600 M-1s-1 by the action of a lysine side chain in a hydrophobic environment [135].
An even bigger challenge was the computational design of a Diels-Alderase as it catalyzes the intermolecular C-C bond formation of two substrates [136]. Of the initially 1019 variants calculated by QM simulations, 106 could be fitted into protein scaffolds and after further optimization finally 84 designs were chosen for biochemical characterization, of which 50 were expressed as soluble proteins but only two showed the desired activity. Although the turnover rate is very low (2 s-1), one variant is highly substrate- and stereospecific forming >97% of the stereoisomer (out of 8 possible) of the substrate it was designed for. An interesting approach was chosen for further improvement of this designed Diels-Alderase variant by challenging the players of the online computer game Foldit, which was created by the Baker Lab to help to predict protein structures [137], to remodel the active-site loops to enable additional interaction with the substrate [138].
The most recent de novo design is the introduction of a catalytic Cys-His dyad and oxyanion holes for ester hydrolysis into different protein folds [139]. The best design so far, which was further improved by site-specific mutagenesis reached a kcat/KM value of 405 M-1s-1, which is still lower than catalytic antibodies. Again structural analysis revealed bottlenecks, which will help to improve subsequent designs.
Artificial metalloenzymes
Artificial metalloenzymes are thought as a bridge between biocatalysis and transition-metal complexes by incorporating the catalytically active transition metal complex in the protein scaffold enabling a high activity and selectivity [105, 140]. There are several approaches how this can be achieved, which can be divided in two main categories: non-covalent anchoring, where either the affinity of a protein for a transition metal is used (dative anchoring) or high-affinity protein substrate interactions are employed (supramolecular anchoring), and covalent modification (for a review see [141]). The range of metal ions that can be incorporated in the active site increases the range of chemical transformations catalyzed by the enzyme. Some enzymes already display promiscuous activity due to different metal ions present in the active site (e.g. [142, 143]). In some cases the intrinsic metal ions of natural metalloproteins were successfully exchanged to different metal ions, thereby altering the reaction catalyzed by the enzyme. Carbonic anhydrase II is a zinc metalloenzyme with Zn2+ interacting with three His residues. Its physiological role is the catalysis of the reversible hydration of carbon dioxide with a kcat/KM value close to the diffusion limit. However, carbonic anhydrase II shows also promiscuous esterase activity [144]. The replacement of the Zn2+ in human carbonic anhydrase by Mn2+ resulted in a peroxidase, which catalyzes the efficient oxidation of o-dianisidine (kcat/KM = 1.4x106) in the presence of bicarbonate and hydrogen peroxide (H2O2), and also epoxidation of olefins, however with only low to moderate enantioselectivity [145, 146]. The same group also showed that rhodium-substituted carbonic anhydrase induced stereoselective hydrogenation of stilbene favoring cis- over trans-stilbene by about 20:1 [147]. It also catalyzes the hydroformylation of olefins with a regioselectivity of 8.4 for linear over branched aldehyde product, which is in opposite to uncomplexed Rh, which results in mainly branched product [148]. Exchange of the metal ion in the Zn2+-dependent β-lactamase from Stenotrophomonas maltophilia to Cu2+ resulted in 10-fold lower lactamase activity and a promiscuous new oxidase activity towards catechol. This oxidase activity was further improved by site-specific mutagenesis to stabilize the coordination of the Cu2+ ion in the second metal binding site [149].
Another option is the creation of a novel metal binding site in a host protein by introduction of coordinating amino acids at geometrically appropriate positions of the protein. Reetz and his group introduced two His residues in the thermostable protein tHisF, the synthase subunit of an imidazole glycerol phosphate synthase, from Thermotoga maritima to create a 2-His-1-carboxylate motif for the binding of Cu2+ [150]. They showed that the enantioselectivity of a model Diels-Alder cycloaddition was enhanced with the protein-Cu2+ catalyst compared to Cu2+ alone. A similar approach was followed by Lu and coworkers, who introduced three histidines and one glutamate or two histidines and one glutamate to create a new non-heme iron center in myoglobin near the unchanged heme cofactor to design a nitric oxide reductase (NOR) [152–154], which should help to understand the reaction mechanism of naturally occurring NORs. Both enzymes converted NO to N2O. Myoglobin was found to display weak nitrite reductase (NiR) activity. The X-ray structure of NiR identified the location of two His and one Tyr residues, which are essential for NiR activity. As for NOR, suitable target residues for mutagenesis were identified in myoglobin by molecular modeling in combination with molecular dynamics simulations giving an insight into the nitrite binding by calculation of theoretical KM-values [154]. However, so far experimental data are missing.
In yet a different strategy mononuclear zinc metalloenzymes were computationally redesigned to catalyze the hydrolysis of organophosphates by changing active site amino acids while maintaining the coordination geometry around the metal ion [155]. Of twelve designs a redesigned adenosine deaminase hydrolyzed the achiral organophosphate diethyl 7-hydroxycoumarinyl phosphate (DECP) and was further improved by directed evolution (kcat/KM values up to 104 M-1s-1). It also hydrolyzed the coumarinyl analog of the nerve agent cyclosarin with a preference for the RP isomer.
Conclusion
This review describes recent advances in enzyme engineering focusing on rational approaches for the creation of biocatalysts with improved activity, enantioselectivity, substrate range, stability and also new catalytic functions. It is highlighted that in many cases protein structures helped to understand the basis of the enzyme's reaction mechanism and allowed the successful fine-tuning of the active site by rational design by only a few specific mutations thereby providing a strong alternative to extensive screening of large libraries with directed evolution approaches.
However, despite the availability of a fast-growing number of protein structures and highly sophisticated computational algorithms pure rational design is still limited by (i) our incomplete understanding of the enzyme functions and protein folding, (ii) the flexibility of the proteins and conformational changes upon e.g. substrate binding, and thus (iii) our still limited understanding of protein dynamics [122], (iv) the sensitivity of the enzymatic reaction to small changes in distances and geometry of the substrate in relation to the active site amino acids and to the influence of active site water molecules [139, 123], (v) the adverse effect of some mutations on the expression and stability of the protein (e.g. [156, 157]), (vi) the positive effect of apparently unrelated distant mutations as well as correlated mutations, which are at the moment difficult to predict [158, 40, 17, 24] and (vii) the enormous computational capacity needed for processing large data volumes.
Thus, pure computational design of de novo enzymes is still in its infancies and the tailored biocatalysts show just small, but promising activities. However, the probability of the success of solely random strategies is also low and requires high screening effort. This review provides several examples, in which the combination of rational design with subsequent directed evolution led to the most promising de novo enzymes.
In summary, there exists no general strategy how to proceed in the engineering of a certain enzyme. It strongly depends on the reaction and enzyme of interest, the biochemical and structural data available, the structural bioinformatics expertise and computational equipment, and the library screening capabilities to mention just a few. Currently, computational enzyme design has fundamentally changed the way biocatalysts can be altered, but it cannot yet replace directed evolution as a method of choice for protein engineering. On the contrary, the two approaches are complementary and should be combined for reaching the most promising results either in semi-rational approaches or using them subsequently.
Acknowledgements
This work has been supported by the Federal Ministry of Economy, Family and Youth (BMWFJ), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and ZIT - Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG.
Competing Interests
The authors have declared that no competing interests exist.
References
- 1.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, et al. (2012) Engineering the third wave of biocatalysis. Nature 485: 185–194 [DOI] [PubMed] [Google Scholar]
- 2.Drauz K, Gröger H, May O (2012). Enzyme Catalysis in Organic Synthesis. Weinheim: Wiley-VCH Verlag GmbH & Co [Google Scholar]
- 3.Behrens GA, Hummel A, Padhi SK, Schätzle S, Bornscheuer UT (2011) Discovery and Protein Engineering of Biocatalysts for Organic Synthesis. Adv Synth Catal 353: 2191–2215 [Google Scholar]
- 4.Bommarius AS, Blum JK, Abrahamson MJ (2011) Status of protein engineering for biocatalysts: how to design an industrially useful biocatalyst. Curr Opin Chem Biol 15: 194–200 [DOI] [PubMed] [Google Scholar]
- 5.Brustad EM, Arnold FH (2011) Optimizing non-natural protein function with directed evolution. Curr Opin Chem Biol 15: 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlt JA, Babbitt PC (2009) Enzyme (re)design: lessons from natural evolution and computation. Curr Opin Chem Biol 13: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz S (2010) Beyond directed evolution - semi-rational protein engineering and design. Curr Opin Biotechnol 21: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reetz MT (2011) Laboratory Evolution of Stereoselective Enzymes: A Prolific Source of Catalysts for Asymmetric Reactions. Angew Chem 50: 138–174 [DOI] [PubMed] [Google Scholar]
- 9.Strohmeier G, Pichler H, May O, Gruber-Khadjawi M (2011) Application of Designed Enzymes in Organic Synthesis. Chem Rev 111: 4141–4164 [DOI] [PubMed] [Google Scholar]
- 10.Chica RA, Doucet N, Pelletier JN (2005) Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr Opin Biotechnol 16: 378–384 [DOI] [PubMed] [Google Scholar]
- 11.Abrahamson MJ, Vazquez-Figueroa E, Woodall NB, Moore JC, Bommarius AS (2012) Development of an Amine Dehydrogenase for Synthesis of Chiral Amines. Angew Chem 51: 3969–3972 [DOI] [PubMed] [Google Scholar]
- 12.Dudek HM, de Gonzalo G, Pazmino DET, Stepniak P, Wyrwicz LS, et al. (2011) Mapping the Substrate Binding Site of Phenylacetone Monooxygenase from Thermobifida fusca by Mutational Analysis. Appl Environ Microbiol 77: 5730–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reetz MT, Wang L, Bocola M (2006) Directed Evolution of Enantioselective Enzymes: Iterative Cycles of CASTing for Probing Protein-Sequence Space. Angew Chem 45: 1236–1241 [DOI] [PubMed] [Google Scholar]
- 14.Allen BD, Nisthal A, Mayo SL (2010) Experimental library screening demonstrates the successful application of computational protein design to large structural ensembles. Proc Natl Acad Sci USA 107: 19838–19843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen TS, Keating AE (2012) Designing specific protein-protein interactions using computation, experimental library screening, or integrated methods. Protein Sci 21: 949–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lappe M, Bagler G, Filippis I, Stehr H, Duarte JM, et al. (2009) Designing evolvable libraries using multi-body potentials. Curr Opin Biotechnol 20: 437–446 [DOI] [PubMed] [Google Scholar]
- 17.Lippow SM, Moon TS, Basu S, Yoon SH, Li X, et al. (2010) Engineering Enzyme Specificity Using Computational Design of a Defined-Sequence Library. Chem Biol 17: 1306–1315 [DOI] [PubMed] [Google Scholar]
- 18.Treynor TP, Vizcarra CL, Nedelcu D, Mayo SL (2007) Computationally designed libraries of fluorescent proteins evaluated by preservation and diversity of function. Proc Natl Acad Sci USA 104: 48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann M, Wyss M (2001) Engineering proteins for thermostability: the use of sequence alignments versus rational design and directed evolution. Curr Opin Biotechnol 12: 371–375 [DOI] [PubMed] [Google Scholar]
- 20.Lehmann M, Loch C, Middendorf A, Studer D, Lassen S, et al. (2002) The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng 15: 403–411 [DOI] [PubMed] [Google Scholar]
- 21.Anbar M, Gul O, Lamed R, Sezerman UO, Bayer EA (2012) Improved thermostability of Clostridium thermocellum endoglucanase Cel8A using consensus-guided mutagenesis. Appl Environ Microbiol 78: 3458–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muslin E, Clark S, Henson C (2002) The effect of proline insertions on the thermostability of a barley α-glucosidase. Protein Eng 15: 29–33 [DOI] [PubMed] [Google Scholar]
- 23.Tian J, Wang P, Gao S, Chu X, Wu N, et al. (2010) Enhanced thermostability of methyl parathion hydrolase from Ochrobactrum sp. M231 by rational engineering of a glycine to proline mutation. FEBS Journal 277: 4901–4908 [DOI] [PubMed] [Google Scholar]
- 24.Morley KL, Kazlauskas RJ (2005) Improving enzyme properties: when are closer mutations better?. Trends Biotechnol 23: 231–237 [DOI] [PubMed] [Google Scholar]
- 25.Polizzi K, Chaparro-Riggers J, Vazquez-Figueroa E, Bommarius AS (2006) Structure-guided consensus approach to create a more thermostable penicillin G acylase. Biotechnol J 1: 531–536 [DOI] [PubMed] [Google Scholar]
- 26.Steipe B, Schiller B, Pückthun A, Steinbacher S (1994) Sequence statistics reliably predict stabilizing mutations in a protein domain. J Mol Biol 240: 188–192 [DOI] [PubMed] [Google Scholar]
- 27.Vazquez-Figueroa E, Chaparro-Riggers J, Bommarius AS (2007) Development of a Thermostable Glucose Dehydrogenase by a Structure-Guided Consensus Concept. Chembiochem 8: 2295–2301 [DOI] [PubMed] [Google Scholar]
- 28.Vazquez-Figueroa E, Yeh V, Broering J, Chaparro-Riggers J, Bommarius A (2008) Thermostable variants constructed via the structure-guided consensus method also show increased stability in salts solutions and homogeneous aqueous-organic media. Protein Eng Des Sel 21: 673–680 [DOI] [PubMed] [Google Scholar]
- 29.Blum JK, Ricketts MD, Bommarius A (2012) Improved thermostability of AEH by combining B-FIT analysis and structure-guided consensus method. J Biotechnol 160: 214–212 [DOI] [PubMed] [Google Scholar]
- 30.Augustyniak W, Brzezinska A, Pijning T, Wienk H, Boelens R, et al. (2012) Biophysical characterization of mutants of Bacillus subtilis lipase evolved for thermostability: Factors contributing to increased activity retention. Protein Sci 21: 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parthasarathy S, Murthy M (2000) Protein thermal stability: insights from atomic displacement parameters (B values). Protein Eng 13: 9–13 [DOI] [PubMed] [Google Scholar]
- 32.Reetz MT, Carballeira J, Vogel A (2006) Iterative Saturation Mutagenesis on the Basis of B Factors as a Strategy for Increasing Protein Thermostability. Angew Chem 118: 7909–7915 [DOI] [PubMed] [Google Scholar]
- 33.Reetz MT, Soni P, Fernandez L, Gumulya Y, Carballeira JD (2010) Increasing the stability of an enzyme toward hostile organic solvents by directed evolution based on iterative saturation mutagenesis using the B-FIT method. Chem Commun 46: 8657–8658 [DOI] [PubMed] [Google Scholar]
- 34.Cole MF, Gaucher EA (2011) Utilizing natural diversity to evolve protein function: applications towards thermostability. Curr Opin Chem Biol 15: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaucher EA, Govindarajan S, Ganesh OK (2008) Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451: 704–708 [DOI] [PubMed] [Google Scholar]
- 36.Bergthorsson U, Andersson DI, Roth JR (2007) Ohno's dilemma: Evolution of new genes under continuous selection. Proc Natl Acad Sci USA 104: 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe K, Ohkuri T, Yokobori S, Yamagishi A (2006) Designing thermostable proteins: Ancestral mutants of 3-isopropylmalate dehydrogenase designed by using a phylogenetic tree. J Mol Biol 355: 664–674 [DOI] [PubMed] [Google Scholar]
- 38.Yamashiro K, Yokobori S, Koikeda S, Yamagishi A (2010) Improvement of Bacillus circulans beta-amylase activity attained using the ancestral mutation method. Prot Engin Des Sel 23: 519–528 [DOI] [PubMed] [Google Scholar]
- 39.Kourist R, Jochens H, Bartsch S, Kuipers R, Padhi S, et al. (2010) The α/β-Hydrolase Fold 3DM Database (ABHDB) as a Tool for Protein Engineering. Chembiochem 11: 1635–1643 [DOI] [PubMed] [Google Scholar]
- 40.Kuipers RK, Joosten H, Verwiel E, Paans S, Akerboom J, et al. (2009) Correlated mutation analyses on super-family alignments reveal functionally important residues. Proteins 76: 608–616 [DOI] [PubMed] [Google Scholar]
- 41.Kuipers RK, Joosten HJ, van Berkel WJH, Leferink NGH, Rooijen E, et al. (2010) 3DM: Systematic analysis of heterogeneous superfamily data to discover protein functionalities. Proteins 78: 2101–2113 [DOI] [PubMed] [Google Scholar]
- 42.Jochens H, Bornscheuer UT (2010) Natural Diversity to Guide Focused Directed Evolution. Chembiochem 11: 1861–1866 [DOI] [PubMed] [Google Scholar]
- 43.Jochens H, Aerts D, Bornscheuer U (2010) Thermostabilization of an esterase by alignment-guided focussed directed evolution. Protein Eng Des Sel 23: 903–909 [DOI] [PubMed] [Google Scholar]
- 44.Bassegoda A, Nguyen GS, Schmidt M, Kourist R, Diaz P, et al. (2010) Rational Protein Design of Paenibacillus barcinonensis Esterase EstA for Kinetic Resolution of Tertiary Alcohols. ChemCatChem 2: 962–967 [Google Scholar]
- 45.Cerdobbel A, De Winter K, Aerts D, Kuipers R, Joosten H, et al. (2011) Increasing the thermostability of sucrose phosphorylase by a combination of sequence- and structure-based mutagenesis. Protein Eng Des Sel 24: 829–834 [DOI] [PubMed] [Google Scholar]
- 46.Leferink NGH, Fraaije MW, Joosten HJ, Schaap PJ, Mattevi A, et al. (2009) Identification of a Gatekeeper Residue That Prevents Dehydrogenases from Acting as Oxidases. J Biol Chem 284: 4392–4397 [DOI] [PubMed] [Google Scholar]
- 47.Tengg M, Stecher H, Remler P, Eiteljörg I, Schwab H (2012) Molecular Characterization of the C-Methyltransferase NovO of Streptomyces spheroides, a valuable enzyme for performing Friedel-Crafts alkylation. J Mol Catal B 84: 2–8 [Google Scholar]
- 48.Meyer MM, Silberg JJ, Voigt CA, Endelman JB, Mayo SL, et al. (2003) Library analysis of SCHEMA-guided protein recombination. Protein Sci 12: 1686–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer MM, Hochrein L, Arnold FH (2006) Structure-guided SCHEMA recombination of distantly related beta-lactamases. Protein Eng Des Sel 19: 563–570 [DOI] [PubMed] [Google Scholar]
- 50.Otey CR, Landwehr M, Endelman JB, Hiraga K, Bloom JD, et al. (2006) Structure-guided recombination creates an artificial family of cytochromes P450. PLoS Biology 4: 789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinzelman P, Snow CD, Smith MA, Yu XL, Kannan A, et al. (2009) SCHEMA Recombination of a Fungal Cellulase Uncovers a Single Mutation That Contributes Markedly to Stability. J Biol Chem 284: 26229–26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinzelman P, Komor R, Kanaan A, Romero P, Yu XL, et al. (2010) Efficient screening of fungal cellobiohydrolase class I enzymes for thermostabilizing sequence blocks by SCHEMA structure-guided recombination. Protein Eng Des Sel 23: 871–880 [DOI] [PubMed] [Google Scholar]
- 53.Li YG, Drummond DA, Sawayama AM, Snow CD, Bloom JD, et al. (2007) A diverse family of thermostable cytochrome P450s created by recombination of stabilizing fragments. Nat Biotechnol 25: 1051–1056 [DOI] [PubMed] [Google Scholar]
- 54.Landwehr M, Carbone M, Otey CR, Li YG, Arnold FH (2007) Diversification of catalytic function in a synthetic family of chimeric cytochrome P450s. Chem Biol 14: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Beek HL, de Gonzalo G, Fraaije MW (2012) Blending Baeyer-Villiger monooxygenases: using a robust BVMO as a scaffold for creating chimeric enzymes with novel catalytic properties. Chem Commun 48: 3288–3290 [DOI] [PubMed] [Google Scholar]
- 56.Verschueren E, Vanhee P, van der Sloot AM, Serrano L, Rousseau F, et al. (2011) Protein design with fragment databases. Curr Opin Struct Biol 21: 452–459 [DOI] [PubMed] [Google Scholar]
- 57.Svedendahl Humble M, Berglund P (2011) Biocatalytic Promiscuity. Eur J Org Chem 2: 3391–3401 [Google Scholar]
- 58.Bornscheuer UT, Kazlauskas RJ (2004) Catalytic promiscuity in biocatalysis: Using old enzymes to form new bonds and follow new pathways. Angew Chem 43: 6032–6040 [DOI] [PubMed] [Google Scholar]
- 59.Hult K, Berglund P (2007) Enzyme promiscuity: mechanism and applications. Trends Biotechnol 25: 231–238 [DOI] [PubMed] [Google Scholar]
- 60.Kazlauskas RJ (2005) Enhancing catalytic promiscuity for biocatalysis. Curr Opin Chem Biol 9: 195–201 [DOI] [PubMed] [Google Scholar]
- 61.Terao Y, Miyamoto K, Ohta H (2006) Introduction of single mutation changes arylmalonate decarboxylase to racemase. Chem Commun 34: 3600–3602 [DOI] [PubMed] [Google Scholar]
- 62.Meyer D, Walter L, Kolter G, Pohl M, Müller M, et al. (2011) Conversion of Pyruvate Decarboxylase into an Enantioselective Carboligase with Biosynthetic Potential. J Am Chem Soc 133: 3609–3616 [DOI] [PubMed] [Google Scholar]
- 63.Seebeck FP, Hilvert D (2003) Conversion of a PLP-dependent racemase into an aldolase by a single active site mutation. J Am Chem Soc 125: 10158–10159 [DOI] [PubMed] [Google Scholar]
- 64.Seebeck FP, Guainazzi A, Amoreira C, Baldridge KK, Hilvert D (2006) Stereoselectivity and expanded substrate scope of an engineered PLP-dependent aldolase. Angew Chem 45: 6824–6826 [DOI] [PubMed] [Google Scholar]
- 65.Toscano MD, Müller MM, Hilvert D (2007) Enhancing activity and controlling stereoselectivity in a designed PLP-dependent aldolase. Angew Chem 46: 4468–4470 [DOI] [PubMed] [Google Scholar]
- 66.Yow GY, Watanabe A, Yoshimura T, Esaki N (2003) Conversion of the catalytic specificity of alanine racemase to a D-amino acid aminotransferase activity by a double active-site mutation. J Mol Catal B 23: 311–319 [Google Scholar]
- 67.Jochens H, Hesseler M, Stiba K, Padhi SK, Kazlauskas RJ, et al. (2011) Protein Engineering of alpha/beta-Hydrolase Fold Enzymes. Chembiochem 12: 1508–1517 [DOI] [PubMed] [Google Scholar]
- 68.Bernhardt P, Halt K, Kazlauskas RJ (2005) Molecular basis of perhydrolase activity in serine hydrolases. Angew Chem 44: 2742–2746 [DOI] [PubMed] [Google Scholar]
- 69.Yin DL, Kazlauskas RJ (2012) Revised Molecular Basis of the Promiscuous Carboxylic Acid Perhydrolase Activity in Serine Hydrolases. Chem Eur J 18: 8130–8139 [DOI] [PubMed] [Google Scholar]
- 70.Feichtenhofer S, Höffken WW, Schwab H (2009) Conversion of Esterase EstC from Burkholderia gladioli into a Hydroxynitrile lyase. Poster at Enzyme Engineering XX, Groningen, The Netherlands [Google Scholar]
- 71.Padhi S, Fujii R, Legatt G, Fossum S, Berchtold R, et al. (2010) Switching from an Esterase to a Hydroxynitrile Lyase Mechanism Requires Only Two Amino Acid Substitutions. Chem Biol 17: 863–871 [DOI] [PubMed] [Google Scholar]
- 72.Jochens H, Stiba K, Savile C, Fujii R, Yu J, et al. (2009) Converting an Esterase into an Epoxide Hydrolase. Angew Chem 48: 3532–3535 [DOI] [PubMed] [Google Scholar]
- 73.Kourist R, Bartsch S, Fransson L, Hult K, Bornscheuer UT (2008) Understanding promiscuous amidase activity of an esterase from Bacillus subtilis. Chembiochem 9: 67–69 [DOI] [PubMed] [Google Scholar]
- 74.Torres LL, Schliessmann A, Schmidt M, Silva-Martin N, Hermoso JA, et al. (2012) Promiscuous enantioselective (-)-γ-lactamase activity in the Pseudomonas fluorescens esterase I. Org Biomol Chem 10: 3388–3392 [DOI] [PubMed] [Google Scholar]
- 75.Syren PO, Hult K (2011) Amidases Have a Hydrogen Bond that Facilitates Nitrogen Inversion, but Esterases Have Not. ChemCatChem 3: 853–860 [Google Scholar]
- 76.Branneby C, Carlqvist P, Hult K, Brinck T, Berglund P (2003) Rational redesign of a lipase to an aldolase. Biochemistry 42: 8633 [Google Scholar]
- 77.Carlqvist P, Svedendahl M, Branneby C, Hult K, Brinck T, et al. (2005) Exploring the active-site of a rationally redesigned lipase for catalysis of Michael-type additions. Chembiochem 6: 331–336 [DOI] [PubMed] [Google Scholar]
- 78.Svedendahl M, Hult K, Berglund P (2005) Fast carbon-carbon bond formation by a promiscuous lipase. J Am Chem Soc 127: 17988–17989 [DOI] [PubMed] [Google Scholar]
- 79.Svedendahl M, Jovanovic B, Fransson L, Berglund P (2009) Suppressed Native Hydrolytic Activity of a Lipase to Reveal Promiscuous Michael Addition Activity in Water. ChemCatChem 1: 252–258 [Google Scholar]
- 80.Strohmeier G, Sovic T, Steinkellner G, Hartner F, Andryushkova A, et al. (2009) Investigation of lipase-catalyzed Michael-type carbon-carbon bond formations. Tetrahedron 65: 5663–5668 [Google Scholar]
- 81.Mutti FG, Lara M, Kroutil M, Kroutil W (2010) Ostensible Enzyme Promiscuity: Alkene Cleavage by Peroxidases. Chem Eur J 16: 14142–14148 [DOI] [PubMed] [Google Scholar]
- 82.Hickel A, Heinrich G, Schwab H, Griengl H (1997) Screening for hydroxynitrile lyases in plants. Biotechnol Techn 11: 55–58 [Google Scholar]
- 83.Evitt AS, Bornscheuer UT (2011) Lipase CAL-B does not catalyze a promiscuous decarboxylative aldol addition or Knoevenagel reaction. Green Chem 13: 1141–1142 [Google Scholar]
- 84.Weis R, Gaisberger R, Skranc W, Gruber K, Glieder A (2005) Carving the active site of almond R-HNL for increased enantioselectivity. Angew Chem 44: 4700–4704 [DOI] [PubMed] [Google Scholar]
- 85.Bühler H, Effenberger F, Förster S, Roos J, Wajant H (2003) Substrate specificity of mutants of the hydroxynitrile lyase from Manihot esculenta. Chembiochem 4: 211–216 [DOI] [PubMed] [Google Scholar]
- 86.Lauble H, Miehlich B, Förster S, Kobler C, Wajant H, et al. (2002) Structure determinants of substrate specificity of hydroxynitrile lyase from Manihot esculenta. Protein Sci 11: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winkler M, Glieder A, Steiner K (2012) Hydroxynitrile lyases: From Nature to Application In: Comprehensive Chirality. Elsevier: In Press [Google Scholar]
- 87b.Dadashipour M, Asano Y, et al. (2011) Hydroxynitrile Lyases: Insights into Biochemistry, Discovery and Engineering. ACS Catalysis 1: 1121–1149 [Google Scholar]
- 88.Pavlova M, Klvana M, Prokop Z, Chaloupkova R, Banas P (2009) Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat Chem Biol 5: 727–733 [DOI] [PubMed] [Google Scholar]
- 89.Ema T, Nakano Y, Yoshida D, Kamata S, Sakai T (2012) Redesign of enzyme for improving catalytic activity and enantioselectivity toward poor substrates: manipulation of the transition state. Org Biomol Chem 10: 6299–6308 [DOI] [PubMed] [Google Scholar]
- 90.Liu L, Murphy P, Baker D, Lutz S, et al. (2010) Computational design of orthogonal nucleoside kinases. Chem Commun 46: 8803–8805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, et al. (2010) Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science 329: 309–313 [DOI] [PubMed] [Google Scholar]
- 92.Cho BK, Park HY, Seo JH, Kim JH, Kang TJ (2008) Redesigning the substrate specificity of omega-aminotransferase for the kinetic resolution of aliphatic chiral amines. Biotechnol Bioengin 99: 275–284 [DOI] [PubMed] [Google Scholar]
- 93.Svedendahl Humble M, Cassimjee KE, Abedi V, Federsel H, Berglund P (2012) Key Amino Acid Residues for Reversed or Improved Enantiospecificity of an ω-Transaminase. ChemCatChem 4: 1167–1172 [Google Scholar]
- 94.Svedendahl M, Branneby C, Lindberg L, Berglund P (2010) Reversed Enantiopreference of an omega-Transaminase by a Single-Point Mutation. ChemCatChem 2: 976–980 [Google Scholar]
- 95.Mathew S, Yun H (2012) Omega-Transaminases for the Production of Optically Pure Amines and Unnatural Amino Acids. ACS Catalysis 2: 993–1001 [Google Scholar]
- 96.Ivancic M, Valinger G, Gruber K, Schwab H (2007) Inverting enantioselectivity of Burkholderia gladioli esterase EstB by directed and designed evolution. J Biotechnol 129: 109–122 [DOI] [PubMed] [Google Scholar]
- 97.Bartsch S, Kourist R, Bornscheuer UT (2008) Complete inversion of enantioselectivity towards acetylated tertiary alcohols by a double mutant of a Bacillus subtilis esterase. Angew Chem 47: 1508–1511 [DOI] [PubMed] [Google Scholar]
- 98.Petschacher B, Nidetzky B (2005) Engineering Candida tenuis xylose reductase for improved utilization of NADH: Antagonistic effects of multiple side chain replacements and performance of site-directed mutants under simulated in vivo conditions. Appl Environ Microbiol 71: 6390–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petschacher B, Nidetzky B (2008) Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microbial Cell Factories 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Woodyer R, van der Donk WA, Zhao HM, et al. (2003) Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design. Biochemistry 42: 11604–11614 [DOI] [PubMed] [Google Scholar]
- 101.Ashida H, Galkin A, Kulakova L, Sawa Y, Nakajima N, et al. (2004) Conversion of cofactor specificities of alanine dehydrogenases by site-directed mutagenesis. J Mol Catal B 30: 173–176 [Google Scholar]
- 102.Dudek HM, Pazmino DET, Rodriguez C, de Gonzalo G, Gotor V (2010) Investigating the coenzyme specificity of phenylacetone monooxygenase from Thermobifida fusca. Appl Microbiol Biotechnol 88: 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park JT, Hirano JI, Thangavel V, Riebel BR, Bommarius AS (2011) NAD(P)H oxidase V from Lactobacillus plantarum (NoxV) displays enhanced operational stability even in absence of reducing agents. J Mol Catal B 71: 159–165 [Google Scholar]
- 104.Dahiyat BI, Mayo SL (1997) De novo protein design: Fully automated sequence selection. Science 278: 82–87 [DOI] [PubMed] [Google Scholar]
- 105.Lu Y, Yeung N, Sieracki N, Marshall N (2009) Design of functional metalloproteins. Nature 460: 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nanda V, Koder RL (2010) Designing artificial enzymes by intuition and computation. Nature Chem 2: 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith BA, Hecht MH (2011) Novel proteins: from fold to function. Curr Opin Chem Biol 15: 421–426 [DOI] [PubMed] [Google Scholar]
- 108.Bolon DN, Mayo SL (2001) Enzyme-like proteins by computational design. Proc Natl Acad Sci USA 98: 14274–14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Damborsky J, Brezovsky J (2009) Computational tools for designing and engineering biocatalysts. Curr Opin Chem Biol 13: 26–34 [DOI] [PubMed] [Google Scholar]
- 110.Pantazes RJ, Grisewood MJ, Maranas CD (2011) Recent advances in computational protein design. Curr Opin Struc Biol 21: 467–472 [DOI] [PubMed] [Google Scholar]
- 111.Suarez M, Jaramillo A, et al. (2009) Challenges in the computational design of proteins. J R Soc Interface 6: S477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, et al. (2006) New algorithms and an in silico benchmark for computational enzyme design. Protein Sci 15: 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leaver-Fay A, Tyka M, Lewis S, Lange O, Thompson J (2011) ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 487: 545–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D (2011) De Novo Enzyme Design Using Rosetta3. PLoS One 6: e19230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nosrati G, Houk K (2012) SABER: A computational method for identifying active sites for new reactions. Protein Sci 21: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang M, Si T, Zhao H (2012) Biocatalyst development by directed evolution. Bioresour Technol 115: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baker D (2010) An exciting but challenging road ahead for computational enzyme design. Protein Sci 19: 1817–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Frushicheva M, Cao J, Chu Z, Warshel A (2010) Exploring challenges in rational enzyme design by simulating the catalysis in artificial kemp eliminase. Proc Natl Acad Sci USA 107: 16869–16874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frushicheva M, Cao J, Warshel A (2011) Challenges and advances in validating enzyme design proposals: The case of the kemp eliminase catalysis. Biochemistry 50: 3849–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kiss G, Röthlisberger D, Baker D, Houk K (2010) Evaluation and ranking of enzyme designs. Protein Sci 19: 1760–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lassila J, Baker D, Herschlag D (2010) Origins of catalysis by computationally designed retroaldolase enzymes. Proc Natl Acad Sci USA 107: 4937–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ruscio JZ, Kohn JE, Ball KA, Head-Gordon T, et al. (2009) The Influence of Protein Dynamics on the Success of Computational Enzyme Design. J Am Chem Soc 131: 14111–14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang L, Althoff EA, Bolduc J, Jiang L, Moody J, et al. (2012) Structural Analyses of Covalent Enzyme-Substrate Analog Complexes Reveal Strengths and Limitations of De Novo Enzyme Design. J Mol Biol 415: 615–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Althoff EA, Wang L, Jiang L, Giger L, Lassilla JK, et al. (2012) Robust design and optimization of retroaldol enzymes. Protein Sci 21: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jiang L, Althoff E, Clemente F, Doyle L, Röthlisberger D, et al. (2008) De Novo Computational Design of Retro-Aldol Enzymes. Science 319: 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Khersonsky O, Röthlisberger D, Dym O, Albeck S, Jackson CJ, et al. (2010) Evolutionary Optimization of Computationally Designed Enzymes: Kemp Eliminases of the KE07 Series. J Mol Biol 396: 1025–1042 [DOI] [PubMed] [Google Scholar]
- 127.Khersonsky O, Röthlisberger D, Wollacott A, Murphy P, Dym O, et al. (2011) Optimization of the In-Silico-Designed Kemp Eliminase KE70 by Computational Design and Directed Evolution. J Mol Biol 407: 391–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Röthlisberger D, Khersonsky O, Wollacott A, Jiang L, DeChancie J, et al. (2008) Kemp elimination catalysts by computational enzyme design. Nature 453: 190–195 [DOI] [PubMed] [Google Scholar]
- 129.Khersonsky O, Kiss G, Röthlisberger D, Dym O, Albeck S (2012) Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc Natl Acad Sci USA 109: 10358–10363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lassila JK, Privett HK, Allen BD, Mayo SL, et al. (2006) Combinatorial methods for small-molecule placement in computational enzyme design. Proc Natl Acad Sci USA 103: 16710–16715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Privett HK, Kiss G, Lee TM, Blomberg R, Chica RA, et al. (2012) Iterative approach to computational enzyme design. Proc Natl Acad Sci USA 109: 3790–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Korendovych IV, Kulp DW, Wu YB, Cheng H, Roder H (2011) Design of a switchable eliminase. Proc Natl Acad Sci USA 108: 6823–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Merski M, Shoichet BK (2012) Engineering a model protein cavity to catalyze the Kemp elimination. Proc Natl Acad Sci USA. In press [DOI] [PMC free article] [PubMed]
- 134.Thorn SN, Daniels RG, Auditor MTM, Hilvert D (1995) Large Rate Accelerations in Antibody Catalysis by Strategic Use of Haptenic Charge. Nature 373: 228–230 [DOI] [PubMed] [Google Scholar]
- 135.Hollfelder F, Kirby AJ, Tawfik DS, et al. (1996) Off-the-shelf proteins that rival tailor-made antibodies as catalysts. Nature 383: 60–63 [DOI] [PubMed] [Google Scholar]
- 136.Siegel J, Zanghellini A, Lovick H, Kiss G, Lambert A, et al. (2010) Computational Design of an Enzyme Catalyst for a Stereoselective Bimolecular Diels-Alder Reaction. Science 329: 309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, et al. (2010) Predicting protein structures with a multiplayer online game. Nature 466: 756–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Eiben C, Siegel J, Bale J, Cooper S, Khatib F, et al. (2012) Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nat Biotechnol 30: 190–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Richter F, Blomberg R, Kuzin A, Tong L, Hilvert D (2012) Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. Protein Sci 21: 81–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rosati F, Roelfes G (2010) Artificial Metalloenzymes. ChemCatChem 2: 916–927 [Google Scholar]
- 141.Deuss P, Den Heeten R, Laan W, Kamer P (2011) Bioinspired Catalyst Design and Artificial Metalloenzymes. Chem Eur J 17: 4680–4698 [DOI] [PubMed] [Google Scholar]
- 142.Leitgeb S, Nidetzky B, et al. (2010) Enzyme Catalytic Promiscuity: The Nonheme Fe2 + Center of β-Diketone-Cleaving Dioxygenase Dke1 Promotes Hydrolysis of Activated Esters. Chembiochem 11: 502–505 [DOI] [PubMed] [Google Scholar]
- 143.Sanchez-Moreno I, Iturrate L, Martin-Hoyos R, Jimeno M, Mena M (2009) From Kinase to Cyclase: An Unusual Example of Catalytic Promiscuity Modulated by Metal Switching. Chembiochem 10: 225–229 [DOI] [PubMed] [Google Scholar]
- 144.Gould SM, Tawfik D (2005) Directed evolution of the promiscuous esterase activity of carbonic anhydrase II. Biochemistry 44: 5444–5452 [DOI] [PubMed] [Google Scholar]
- 145.Fernandez-Gacio A, Codina A, Fastrez J, Riant O, Soumillion P (2006) Transforming Carbonic Anhydrase into Epoxide Synthase by Metal Exchange. Chembiochem 7: 1013–1016 [DOI] [PubMed] [Google Scholar]
- 146.Okrasa K, Kazlauskas R (2006) Manganese-Substituted Carbonic Anhydrase as a New Peroxidase. Chem Eur J 12: 1587–1596 [DOI] [PubMed] [Google Scholar]
- 147.Jing Q, Okrasa K, Kazlauskas R (2009) Stereoselective Hydrogenation of Olefins Using Rhodium-Substituted Carbonic Anhydrase-A New Reductase. Chem Eur J 15: 1370–1376 [DOI] [PubMed] [Google Scholar]
- 148.Jing Q, Kazlauskas R (2010) Regioselective Hydroformylation of Styrene Using Rhodium-Substituted Carbonic Anhydrase. ChemCatChem 2: 953–957 [Google Scholar]
- 149.Fujieda N, Hasegawa A, Ishihama K, Itoh S (2012) Artificial Dicopper Oxidase: Rational Reprogramming of Bacterial Metallo-β-lactamase into a Catechol Oxidase. Chem As J 7: 1203–1207 [DOI] [PubMed] [Google Scholar]
- 150.Podtetenieff J, Taglieber A, Bill E, Reijerse E, Reetz M, et al. (2010) An Artificial Metalloenzyme: Creation of a Designed Copper Binding Site in a Thermostable Protein. Angew Chem 49: 5151–5155 [DOI] [PubMed] [Google Scholar]
- 151.Lin YW, Yeung N, Gao YG, Miner KD, Lei LY, et al. (2010) Introducing a 2-His-1-Glu Nonheme Iron Center into Myoglobin Confers Nitric Oxide Reductase Activity. J Am Chem Soc 132: 9970–9972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin YW, Yeung N, Gao YG, Miner KD, Tian SL, et al. (2010) Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin. Proc Natl Acad Sci USA 107: 8581–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yeung N, Lin Y, Gao Y, Zhao X, Russell B (2009) Rational design of a structural and functional nitric oxide reductase. Nature 462: 1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lin Y, Nie C, Liao L, et al. (2012) Rational design of a nitrite reductase based on myoglobin: a molecular modeling and dynamics simulation study. J Mol Model 18: 4409–4415 [DOI] [PubMed] [Google Scholar]
- 155.Khare S, Kipnis Y, Takeuchi R, Ashani Y, Goldsmith M (2012) Computational redesign of a mononuclear zinc metalloenzyme for organophosphate hydrolysis. Nat Chem Biol 8: 294–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Guo Q, Zhao F, Guo SY, Wang XC (2004) The tryptophane residues of dimeric arginine kinase: roles of Trp-208 and Trp-218 in active site and conformation stability. Biochimie 86: 379–386 [DOI] [PubMed] [Google Scholar]
- 157.Tokuriki N, Stricher F, Serrano L, Tawfik DS (2008) How Protein Stability and New Functions Trade Off. Plos Comp Biol 4: e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kowarsch A, Fuchs A, Frishman D, Pagel P (2010) Correlated Mutations: A Hallmark of Phenotypic Amino Acid Substitutions. Plos Computational Biology 6: e1000923. [DOI] [PMC free article] [PubMed] [Google Scholar]