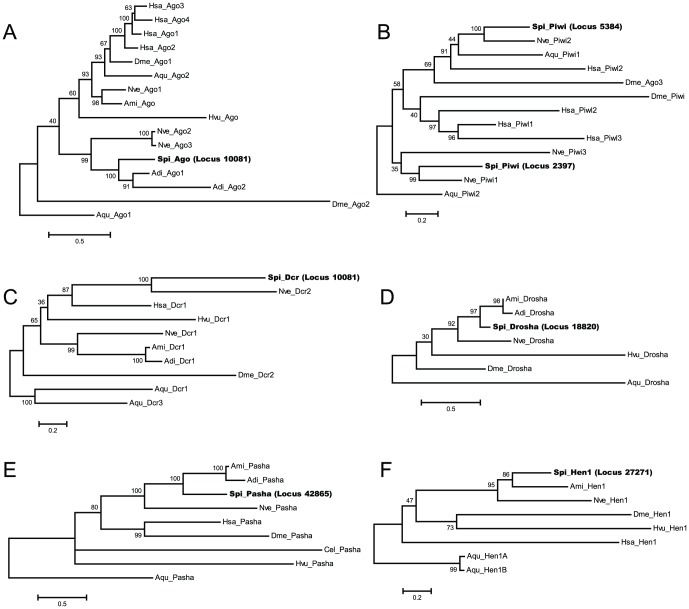
Figure 1. Maximum-likelihood phylogenies of (A) Argonaute, (B) Piwi, (C) Dicer, (D) Drosha, (E) Pasha, and (F) HEN1 proteins.
(A), (C), and (D) were constructed using the amino acid substitution model LG+G, while (B), (E), and (F) were constructed using LG+I+G. Bootstrap support values are indicated above the branches. Abbreviations used in these trees are ‘Adi’: Acropora digitifera; ‘Ami’: Acropora millepora; ‘Aqu’: Amphimedon queenslandica (a sponge); ‘Cel’: C. elegans; ‘Dme’: D. melanogaster; ‘Hsa’: H. sapiens; ‘Hvu’: Hydra vulgaris; ‘Nve’: N. vectensis; and ‘Spi’: S. pistillata.