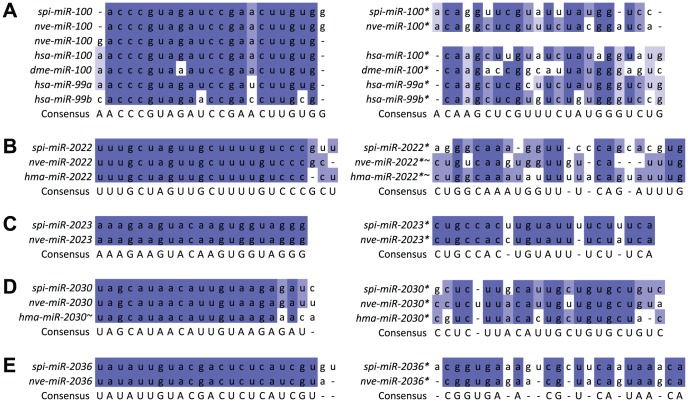
Figure 3. Alignments of predicted S. pistillata miRNAs against (A) members of the miR-100 family; (B) nve- and hma-miR-2022; (C) nve-miR-2023; (D) nve-miR-2030; and (E) nve-miR-2036.
The mature sequences are shown on the left, while star sequences are on the right. Sequences were obtained from miRBase (version 20). The mature hma-miR-2030 aligned best with miR-2030* sequences from N. vectensis and S. pistillata. Sequences marked with a tilde (nve-miR-2022*, hma-miR-2022*, and hma-miR-2030) are miRNAs that we derived based on the alignment of the respective pre-miRNA sequences obtained from miRBase against S. pistillata miRNAs. Bases were coloured to provide visual indication of conservation (dark blue: >80%; blue: >60%; light blue: >40%; uncoloured otherwise). Abbreviations used are ‘dme’: D. melanogaster; ‘hma’: H. magnipapillata; ‘hsa’: H. sapiens; ‘nve’: N. vectensis; and ‘spi’: S. pistillata.