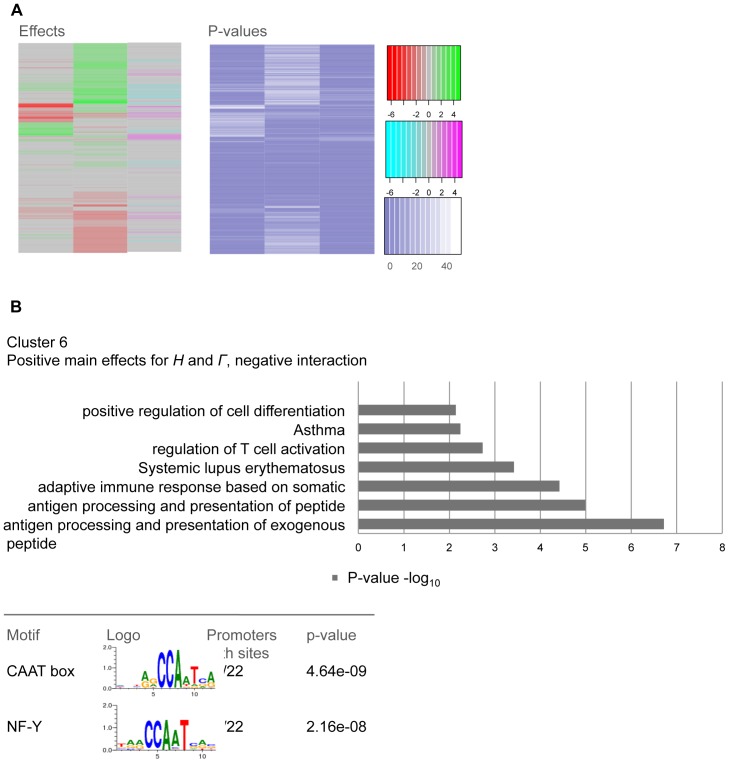
Figure 4. Cluster and pathway analysis.
A: the effect estimates of Model 3 were subjected to a hierarchical cluster analysis. Genes are displayed in the rows, which showed a significant global effect (F-test p-value <0.05 after FDR correction and at least one of the covariates having +/−1.5 fold change). The three columns are the covariates Η, Γ, and Η∶Γ. The column strain shows differences between C57BL/6 and BALB/c, up-regulation shown in red and down-regulation shown in green. The column Γ shows in red up-regulation in BALB/c and in green down-regulation upon IFN-γ stimulation. The third column helps to distinguish alleviating and aggravating effects. Aggravating effects are represented in pink and alleviating effects in turquoise. P-values are plotted separately in a heatmap. The order of the genes is given by the effect estimate clustering. P-values are given in −log10 scale and start from 0 displayed in colors ranging from blue to white. B: The results of a pathway enrichment analysis of cluster 6 as a bar plot. The direction of regulation of the genes of cluster 6 is indicated by the color bar. Gene Ontology ‘Biological Process’ terms and KEGG pathway categories (p<0.01) are sorted from bottom (most significant) to top. To reduce redundancy, similar terms are represented by the most significant and specific term. For complete list of functional annotations see Table S2. The right side shows the results of a TFBS analysis of this gene cluster. The two most significantly represented TFBS are given by the name of the transcription factor, the motif, and the p-value.