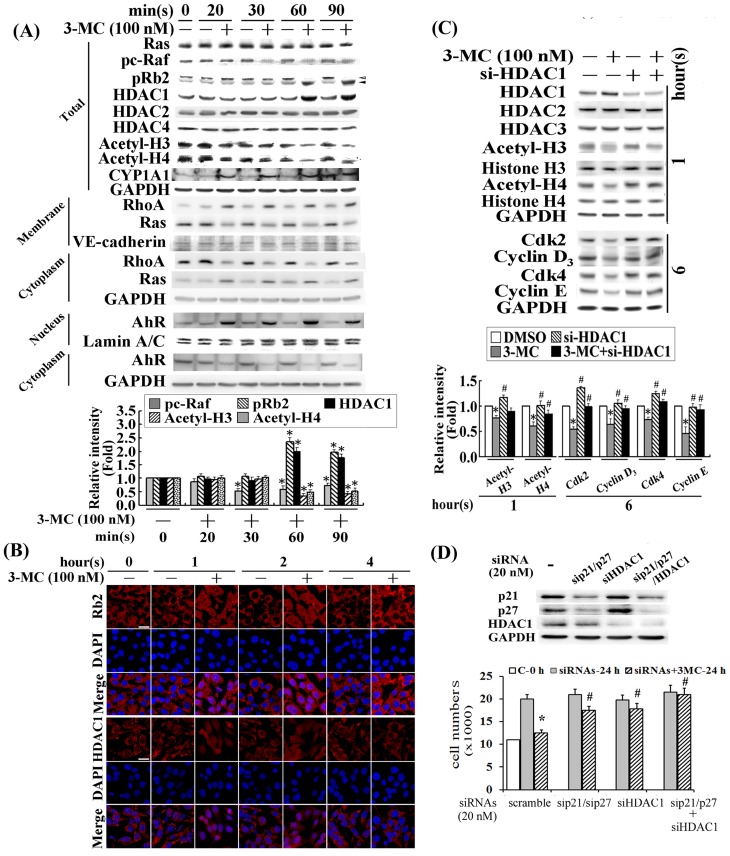
Figure 2. Effects of HDAC1/pRb2 nuclear translocation induced by 3MC in the downregulation of cell-cycle regulatory proteins by histone deacetylation.
(A) Cells were harvested and fractionated from MCVECs treated with 100 nM of 3MC at the indicated times, and were analyzed using Western blot. Anti-GAPDH, anti-Lamin A/C, and anti-VE-cadherin antibodies were used to verify the equivalent loading amounts of the cytosolic, nuclear, and membrane fractions, respectively. The open and closed arrowheads represented the hyper and hypophosphorylated pRb2, respectively. (B) The effect of 3MC on RB2 and HDAC1 subcellular localization was investigated using immunofluorescent chemical staining. Cells cultured in coverslips were challenged by 3MC for 1 h; this was followed by fixation and hybridization using anti-pRb2 and anti-HDAC1 antibodies and then a second antibody conjugated with Texas Red. In Figure 2B, red represents pRb2 or HDAC1-positive staining in the cytosol or nuclei. The identical fields were also stained using DAPI to target the nuclear position. We used 630× magnification (scale bar in white = 50 μm) and recorded micrographs of the representative fields. (C) Cells were transfected with siHDAC1 (20 nM) overnight, prior to treatment with 3MC for 1 or 6 h. The protein levels of acetylated H3/H4, Cdk2/4, and Cyclin D3/E were assayed using Western blot. Membranes were probed using an anti-GAPDH antibody to verify equivalent loading. Three samples were analyzed in each group, and the values reported represent mean ± SEM (*P<0.05 vs. control group; # P<0.05 vs. 3MC treatment alone). (D) Cell numbers were counted for cells with indicated siRNA transfection followed by a 24 h of the 3MC challenge by using a hemo-cytometer. Data are presented as mean ± SEM of 3 independent experiments (*P<0.05 vs. control or siRNA group; # P<0.05 vs. 3MC treatment alone).