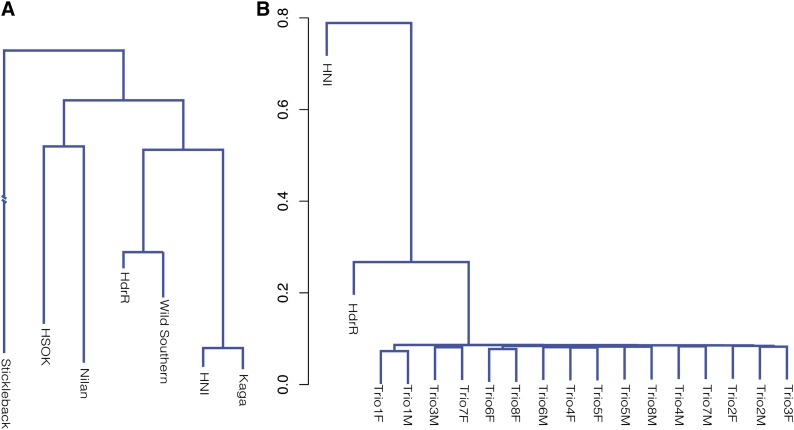
Figure 3.
Phylogenetic relationships of medaka inbred strains and the Kiyosu population. (A) Phylogenetic relationships between the wild Southern population, five inbred strains, and stickleback based on high-throughput genome sequencing. PHYLIP clustering of inbred strains using a neighbor-joining algorithm based on Kimura two-parameter distances is shown. Wild Southern is a synthetic sample based on the sequencing of the Kiyosu population and determining bases using majority vote across 48 haplotypes. Differences between medaka samples and stickleback were too large to estimate distances. (B) Hierarchical clustering with Gower distances of genotypes of Kiyosu population founder samples, and representative Southern and Northern strains.