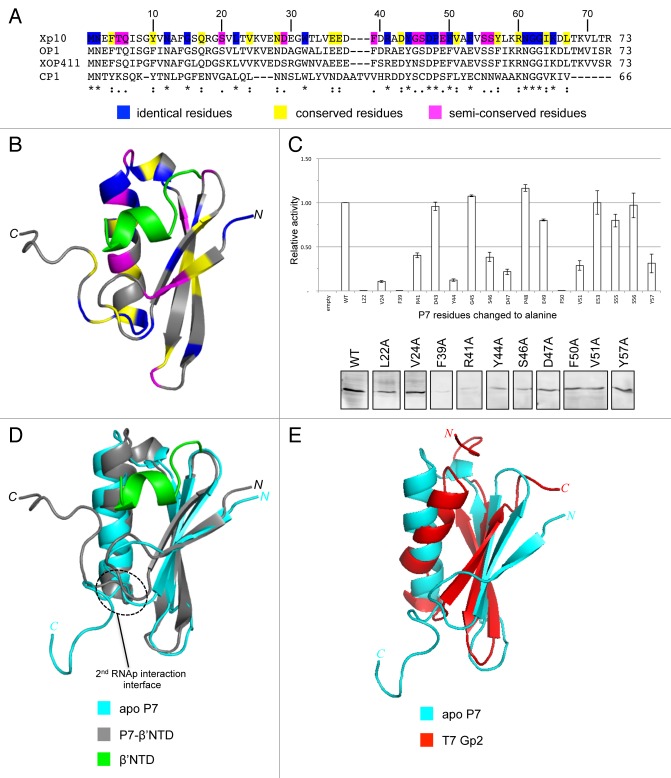
Figure 1. (A) Sequence alignment analysis of Xp10 phage P7-like proteins. The “*,” “:”, and “.” symbols indicate identical, conserved and semi-conserved aa residues, which are also color-coded (in blue, yellow, and magenta, respectively) in the Xp10 phage P7 sequence. (B) Ribbon representation of P7-β’NTD complex.14 The β’NTD is shown in green and the aa residues in P7 that are identical, conserved, and semi-conserved among P7-like proteins are color coded as in (A). (C) Top. Graph showing the ability of P7 mutants harboring alanine substitution at selected aa residues (see text) to interact with the β’NTD relative to the interaction between wild-type P7 and β’NTD. Bottom. Western blot analysis to assess intracellular levels of P7 mutants using polyclonal anti-P7 antibodies. The results rule out the possibility that the failure of the P7 mutants L22A, V24A, D47A, F50A, V51A, and Y57A to interact with the β’NTD is attributable to protein instability under the bacteria two hybrid interaction assay conditions. (D) Overlay of the ribbon representations of apo P7 and P7-β’NTD complex.14 The region on P7 that interacts with the β flap domain is circled. (E) Overlay of the ribbon representations of apo P7 and T7 Gp2.14,15

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.