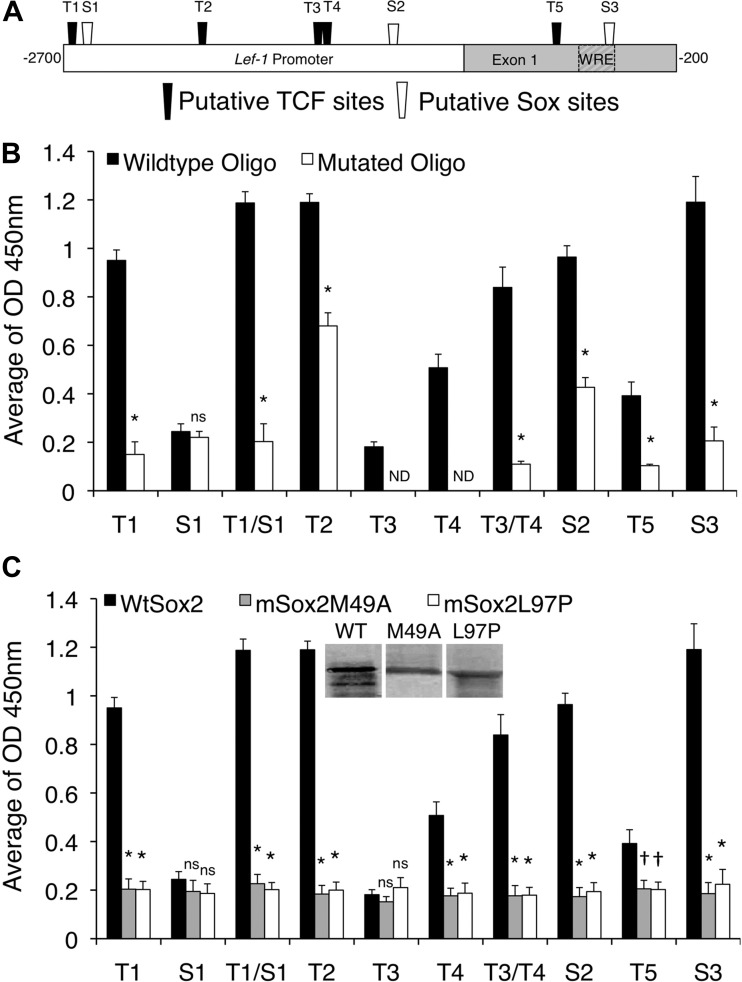
Fig. 6.
Sox2 binds to several putative Sox and TCF family binding sites within the Lef-1 promoter. We analyzed the affinity of bacterially generated Sox2 to eight transcription factor-binding motifs within the Lef-1 promoter using an in vitro NoShift assay. A: schematic of the Lef-1 promoter with five TCF and three Sox family consensus binding sites. Eight wild-type and mutated biotin labeled oligonucleotides were generated to survey the Sox2 binding affinity on these binding motifs (Table 4). B: relative Sox2 binding affinity to wild-type and mutant oligos for the indicated sites. Data represent means ± SE for n = 12 samples. Significant differences using the Mann-Whitney test compared with the wild-type oligo and mutated oligo for each site: *P < 0.001; ns, nonsignificant. C: relative binding of wild type Sox2, Sox2M49A, and Sox2L97P to the wild-type oligo for the indicated sites. Inset shows Western blot of His-tagged Sox2 proteins isolated from bacteria. Data represent means ± SE for n = 12 samples. Significant differences using the Mann-Whitney test compared the wild-type Sox2 vs. each of the mutant Sox2 forms: †P < 0.05; *P < 0.001.