FIG. 1.
(A) Hypothetical protein domain hierarchy. The hierarchy consists of N = 12 sequence subsets (each corresponding to a node; indexed as 1 ≤ z ≤ N) and M = 12 divergent subgroups (each corresponding to a subtree; indexed as 1 ≤ h ≤ M). The tree is colored to highlight subgroup h = 4 (the blue subtree), which is presumed to have diverged most recently from the rest of subgroup h = 2 (maroon nodes). (B) Schematic of a tri-partitioned alignment corresponding to subgroup h = 4 in (A). Such a tri-partition (also called a contrast alignment) is represented mathematically as 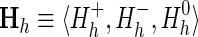 , where each sequence subset (corresponding to each node) is assigned to a foreground partition (
, where each sequence subset (corresponding to each node) is assigned to a foreground partition (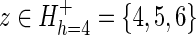 ), a background partition (
), a background partition ( ), or a nonparticipating partition (
), or a nonparticipating partition ( ). The sequences in these partitions are represented by blue, maroon, and gray horizontal bars, respectively. The corresponding nodes in the tree in (A) are colored similarly. Partitioning is based on the conservation of foreground residues (blue vertical bars) that most diverge from (or contrast with) the background residue compositions at those positions (white vertical bars). Red vertical bar heights quantify the degree of divergence. (C) Hypothetical residue sets conserved at discriminating positions in the contrast alignment. The residue set at position j in contrast alignment h is represented mathematically by Ah,j with Ah,j =
). The sequences in these partitions are represented by blue, maroon, and gray horizontal bars, respectively. The corresponding nodes in the tree in (A) are colored similarly. Partitioning is based on the conservation of foreground residues (blue vertical bars) that most diverge from (or contrast with) the background residue compositions at those positions (white vertical bars). Red vertical bar heights quantify the degree of divergence. (C) Hypothetical residue sets conserved at discriminating positions in the contrast alignment. The residue set at position j in contrast alignment h is represented mathematically by Ah,j with Ah,j = 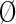 at nondiscriminating positions.
at nondiscriminating positions.

