Abstract
Elevated expression of the anti-apoptotic factor Bcl-2 is believed to be one of the contributing factors to an increased relapse rate associated with multiple cisplatin-resistant cancers. DNA damage-binding protein complex subunit 2 (DDB2) has recently been revealed to play an important role in sensitizing human ovarian cancer cells to cisplatin-induced apoptosis through the down-regulation of Bcl-2, but the underlying molecular mechanism remains poorly defined. Here, we report that DDB2 functions as a transcriptional repressor for Bcl-2 in combination with DDB1. Quantitative ChIP and EMSA analysis revealed that DDB2 binds to a specific cis-acting element at the 5′-end of bcl-2 P1 promoter. Overexpression of DDB2 resulted in marked losses of histone H3K9,14 acetylation along the bcl-2 promoter and enhancer regions, concomitant with a local enrichment of HDAC1 to the bcl-2 P1 core promoter in ovarian cancer cells. Co-immunoprecipitation analysis and in vitro binding assay identified a physical interaction between DDB1 and HDAC1, while downregulation of HDAC1 significantly enhanced bcl-2 promoter activity. Finally, in comparison to wild-type DDB2, mutated DDB2, which is unable to repress Bcl-2 transcription, mediates a compromised apoptosis upon cisplatin treatment. Taken together, our data support a model wherein DDB1 and DDB2 cooperate with each other to repress bcl-2 transcription. DDB2 recognizes and binds to the bcl-2 P1 promoter, and HDAC1 is recruited through the DDB1 subunit associated with DDB2, to deacetylate histone H3K9,14 across bcl-2 regulatory regions, resulting in suppressed bcl-2 transcription. Thus, increasing the expression of DDB complex may provide a molecular strategy for cancer therapy.
Keywords: DNA damage-binding protein (DDB), DDB2, DDB1, bcl-2, Apoptosis, Ovarian cancer, Nucleotide excision repair, transcription, HDAC1
Introduction
Chemoresistance is a challenging barrier for satisfactory cancer treatment outcomes (1). The ability to escape apoptosis, arising from the dysregulated expression of apoptotic factors, appears to be one of the determinants in the acquisition of cisplatin-resistance by ovarian cancers.
The anti-apoptotic factor Bcl-2 is frequently altered at both the transcriptional and translational levels in cisplatin-resistant ovarian and prostate cancers (2, 3). Originally identified in follicular lymphoma (4), human bcl-2 has a complex gene structure and utilizes multiple mechanisms for gene expression in response to environmental and developmental cues. The bcl-2 promoter region spans 3.9 kb in length and contains two promoters. The major promoter P1, located 1,386 to 1,423 bp upstream of the translation start site, is a TATA-less, GC-rich promoter with multiple transcription initiation and Sp1 binding sites. The minor promoter P2, located approximately 1.3 kb downstream of P1, harbors a canonical TATA box, CAAT box, and a simian virus 40 (SV40) decamer/Igoctamer motif (5). Activation of the P1 promoter activity is mediated via the c-AMP response element (CRE) (6, 7), as well as the E2F1 and Sp1 sites (8, 9). The P2 promoter is activated by transcription factors C/EBPα (CCAAT/enhancer binding protein α) and A-Myb (10, 11), but repressed by p53 (12). An enhancer located in the bcl-2 exon II region subjects bcl-2 transcription to further modulation, which is of particular importance in estrogenic regulation of bcl-2 in several hormone-controlled cancers (13). In addition, a long-range chromosomal loop structure formed between the bcl-2 3′ UTR and the promoter region has been revealed recently to be involved in bcl-2 transcription (14). Given the importance of bcl-2 gene in dictating the cell fate, it is not surprising that new regulators are continuously being discovered.
The DNA damage-binding complex (DDB) is composed of two subunits: DDB1 (p127) and DDB2 (p48) (15). Mutations to DDB2 are responsible for the symptoms of Xeroderma Pigmentosum E (XP-E) patients, who exhibit a mild deficiency in global genomic nucleotide excision repair (GGR) and a higher risk of developing skin tumors (16–20). Due to the high affinity of DDB2 for UV-induced DNA lesions, as well as a variety of other damaged DNA forms (20, 21), DDB2 is attributed to be the recognition factor for chromosome lesions during initial steps of GGR (22). The DDB complex is part of a higher-order DDB1-DDB2-Cul4A complex, wherein DDB1 functions as an adaptor bridging DDB2 to the ubiquitin E3 ligase Cul4A during GGR (23–25). Besides its most-studied roles in GGR, DDB2 has come to be recognized as a multifunctional protein, showing involvement in transcriptional regulation through its association with a variety of partners including transcription factor E2F1 (26), transcriptional coactivators CBP/p300 (27), and chromatin remodeling STAGA complex (28).
While the importance of DDB2 in apoptosis has recently received considerable attention, most of the studies have focused on the p53-based apoptosis pathways (29, 30). Considering that p53 is lost or dysfunctional in approximately half of human cancers, it is of special importance to identify new factors that promote apoptosis bypassing the p53-regulated pathways in chemoresistant cancers. We have reported earlier that DDB2 is inversely correlated with Bcl-2 in several cisplatin-resistant human ovarian cancer cell lines, and demonstrated that DDB2 is able to stimulate apoptosis independent of p53 by inhibiting bcl-2 transcription and promoting Bcl-2 degradation via the ubiquitin-proteasome pathway (31). Here, we further explored the underlying mechanism and demonstrated that DDB2 and DDB1 cooperate to repress the bcl-2 promoter activity in human ovarian cancer cells. DDB2 recognizes and binds to the bcl-2 promoter, the associated DDB1 recruits HDAC1 to deacetylate histone H3 along the bcl-2 promoter and enhancer regions.
Material and Methods
Clinical samples
Thirteen frozen tumor tissue samples from ovarian cancer patients were obtained from The Ohio State University Division of Gynecologic Oncology under an approved Institutional Review Board protocol at The Ohio State University. Total protein and RNA were isolated simultaneously from each frozen tissue by Trizol reagent (Invitrogen, Carlsbad, CA).
Cell culture
Cisplatin-sensitive human ovarian cancer cell line A2780 and its derivative cisplatin-resistant subline CP70 were kindly provided by Dr. Paul Modrich (Duke University, Durham, NC). CP70 cells stably transfected with pcDNA3.1-His-DDB2 (CP70-DDB2) were established in our laboratory as described previously (31). These cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. HeLa cells stably transfected with N-terminal FLAG-HA-tagged human DDB2 (HeLa-DDB2) were kindly provided by Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute, Boston, MA). HeLa and HeLa-DDB2 cell lines were cultured in DMEM supplemented with FBS and antibiotics.
Plasmids and cell transfection
The plasmid encoding N-terminal FLAG-tagged wild-type DDB2 (DDB2-WT) and mutant DDB2-K244E have been described previously (32). For transient transfection, the plasmids were delivered into CP70 cells using the Lipofectamine 2000 transfection reagent according to the manufacturer’s instructions (Invitrogen).
Lentivirus assembly and transduction
The HDAC1 shRNA and DDB2 shRNA lentiviruses were generated by co-transfecting HEK293T cells with MISSION shHDAC1 (TRCN0000004816), MISSION shDDB2 (TRCN0000083993) and MISSION Lentiviral Packaging Mix (Sigma, St. Louis, MO) using FuGENE 6 transfection reagents (Promega, Madison, WI) according to the manufacture’s instructions, and further concentrated with Lenti-X Concentrator (Clontech, Mountain View, CA). As a control, the pLKO.1 vector based MISSION Non-target shRNA vector was used to generate the control lentivirus. Target cell lines were transduced with lentiviruses according to the manufactures’ instructions (Sigma).
Dual-luciferase reporter assay
The bcl-2 promoter-luciferase reporter plasmid pGL3 containing the sequence covering both the P1 and P2 promoter, or P2 promoter only (Addgene plasmid 15381 and 15382, respectively) (10), as well as DDB2 expression plasmids were introduced into CP70 cells using the Lipofectamine 2000 transfection reagent (Invitrogen). As an internal control, the pRL4.73 plasmid (Promega), which carries a Renilla luciferase gene, was co-transfected into the cells. The cells were lysed to measure both firefly and renilla luciferase activities by Dual Luciferase Activity Detection System (Promega) 48 h post transfection. Relative luciferase activity was calculated by normalizing the ratio of Firefly/Renilla luciferase activity to that of mock-transfected cells.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was carried out as described previously with a few modifications (33). Briefly, cells were cross-linked with 1% formaldehyde and lysed. Isolated nuclei were digested with Micrococcal nuclease (New England BioLab, Ipswich, MA) and sonicated. Immunoprecipitation (IP) was performed with various ChIP grade antibodies (Table S1). For IP of FLAG-tagged DDB2 from HeLa-DDB2 cells, EZview™ Red ANTI-FLAG® M2 Affinity gel (Sigma) was used. Immunoprecipitated DNA was purified by Phenol/chloroform extraction and quantified by real-time PCR analysis with primer sets corresponding to specific regions of the bcl-2 gene (Table S2).
Quantitative real-time RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen), and the first strand cDNA was generated by the Reverse Transcription System (Promega) in a 20 μl reaction containing 1 μg of total RNA. A 0.5 μl aliquot of cDNA was amplified by Fast SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) in each 20 μl reaction. PCR reactions were run on the ABI 7900 Fast Real-Time PCR system in the OSUCCC Nucleic Acid Core Facility.
Electrophoretic Mobility Shift Assay (EMSA)
The DDB1-DDB2-Cul4A complex (DDB.com) was purified from HeLa-DDB2 cells using the anti-FLAG M2 Magnetic agarose gel (Sigma) as described previously (23). IRDye 700 5′-end-labeled oligos (24 bp) flanking the putative DDB2 binding site BCP1 (underlined) in the bcl-2 P1 promoter (Forward: 5′-IRDye700-ATTAGGAGCTTGAAGCCTTTCAAT-3′; Reverse: 5′-IRDye700-ATTGAAAGGCTTCAAGCTCCTAAT-3′) were synthesized (Integrated DNA Technologies, Coralville, IA), annealed and used as the DNA probe (labeled as Bcl-2 probe). As a positive control, 30 bp-long IRDye 700 5′-end-labeled oligos flanking the reported DDB2 binding site (underlined) in the MnSOD promoter (Forward:5′-IRDye700-GAGGCTAGCCTGCAGCCTCCTTTCTCC-3′; Reverse: 5′-IRDye 700-GGAGAAAGGAGGCTGCAGGCTAGCCTC-3′) were also synthesized (IDT) and annealed (labeled as MnSOD probe). The annealing and binding assay were conducted according to the manufacture’s instructions (Li-Cor, Lincoln, NE). All EMSA assays were conducted in a 20 μl reaction, and the products were resolved in native 4% polyacrylamide gels at 10 V/cm at 4 °C in the dark in 0.5 X Tris borate/EDTA buffer and imaged by Li-Cor Odyssey Imager (Li-Cor).
Immunoblotting and co-IP
Whole cell lysates were prepared by boiling cell pellets for 10 min in lysis buffer (2% SDS, 10% Glycerol, 62 mM Tris-HCl, pH 6.8 and a complete mini-protease inhibitor cocktail [Roche Applied Science]). After protein quantification with Bio-Rad Dc Protein Assay (Bio-Rad Laboratories, Hercules, CA), equal amounts of proteins were loaded, separated on a polyacrylamide gel, and transferred to a nitrocellulose membrane. Protein bands were immuno-detected with appropriate antibodies (Table S1). For co-IP, cells were lysed in cytoplasmic lysis buffer (10 mm Tris-HCl, pH 7.9, 0.34 M sucrose, 1.5 mM MgCl2, 10 mM KCl, 0.5% Nonidet P-40, and protease inhibitors). The isolated nuclei were resuspended in 200 μl of nuclease incubation buffer (50 mM Tris-HCl, pH 7.9, 20 mM NaCl, 1 mM MgCl2) and digested at 37 °C for 30 min with 2 μl of Benzonase (Novagen, Gibbstown, NJ). 200 μl of solubilization buffer (50 mM Tris-HCl, pH 7.9, 300 mM KCl, 1 mM MgCl2, 2% NP40) was added after enzyme digestion and mixed. The chromatin-bound proteins were collected by centrifugation at 18,000 g for 30 min. Equal amount of the solubilized chromatin fraction was immunoprecipitated with anti-FLAG M2 magnetic beads (Sigma), eluted by 2 X SDS loading buffer and followed by immunoblotting analysis.
GST Pull down and binding assay
pGEX4T-GST and pGEX2T-GST-HDAC1 were kindly provided by Dr. Yi Qiu (University of Florida). Bacterially expressed GST or GST-HDAC1 proteins were immobilized on GST-Sepharose 4B beads (GE healthcare, Pataskala, OH), washed, and eluted with 20 mM reduced Glutathione (Sigma) in 50 mM Tris-HCl (pH 8.0). Glutathione was removed by using Amicon® Ultra 10K device and the purified protein was recovered. The protein-protein binding assay was carried out as described (34). Briefly, 2 nmol of purified GST or GST-HDAC1 was mixed with 10 μl 50% GST-Sepharose 4B beads and incubated with gentle rocking for 2 h at 4 °C. The beads were washed with TGEM (1.0) buffer once followed by TGEM (0.1) washing for three times, then incubated with 500 ng purified recombinant FLAG-DDB1 or FLAG-DDB2 protein (Supplementary materials and methods) for another 2 h at 4 °C, washed with TGEM (0.1) buffer for four times, and the bound proteins were released by boiling in SDS sample buffer for 5 min.
Apoptosis analysis by Annexin V staining
Phosphatidylserine exposure on the outer leaflet of the plasma membrane was detected by the Annexin V-FITC apoptosis detection Kit (Cayman Chemical, Ann Arbor, MI) according to the manufacturer’s instructions. Briefly, 1 × 106 cells were pelleted following treatment and washed in PBS. Cells were then resuspended in 500 μl of binding buffer, mixed with Annexin V-FITC and propidium iodide (PI) and incubated at room temperature for 10 min in the dark. The Annexin V-positive cells were analyzed by flow cytometry using BD FACSCalibur flow cytometer (BD, Franklin Lakes, NJ)
Statistical analysis
The correlation between DDB2 and Bcl-2 expression was analyzed with ANOVA. Evaluation of statistical significance was assessed using 2-Sample t-test. Differences were considered to be statistically significance at a value of P < 0.05.
Results
DDB2 is negatively correlated with Bcl-2 in human ovarian tumors
We previously reported that DDB2 expression is inversely correlated with Bcl-2 in several ovarian cancer cell lines (31), this was further confirmed with multiple clones of CP70 cells stably overexpressing DDB2 (Fig. S1A). To evaluate the correlation between DDB2 and Bcl-2 expression in ovarian cancer tissues, a total of thirteen frozen tumor samples were randomly collected from patients diagnosed with late stage ovarian cancer. RNA and protein were isolated out of the same biomass resected from each tissue sample to unambiguously correlate alternations from transcriptome to proteome from sample to sample (35). Consistent with previous observations in human ovarian cancer cell lines, a high Bcl-2/low DDB2 expression pattern was found in the majority of tissue samples (Representative immunoblot images in Fig. S1B). A significant negative correlation (r = −0.754) was revealed by ANOVA analysis between the relative DDB2 and Bcl-2 protein levels in these tissue samples (Fig. 1A). The bcl-2 transcript level is also negatively associated with DDB2 protein level (r = −0.628) (Fig. 1B). However, this correlation is not statistically significant (p=0.052), which is probably due to the relatively small sample size. Collectively, this data suggests that DDB2 negatively regulates Bcl-2 expression in human ovarian tumors.
Figure 1.
DDB2 expression is inversely correlated with Bcl-2 expression in human ovarian tumor tissues. Total protein and RNA were isolated from frozen human ovarian tumor tissues, the protein and mRNA levels of DDB2 and Bcl-2 were analyzed using immunoblotting and real-time RT-PCR, respectively. Correlation between expression of DDB2 protein and Bcl-2 protein (A) or mRNA (B) was determined by Spearman statistical analysis.
DDB2 down-regulates bcl-2 transcription from the upstream P1 promoter
As the regulation of the bcl-2 transcriptional program is a key step in determining the expression and function of the Bcl-2 protein, we next attempted to address how DDB2 exerts its negative control over bcl-2 transcription in ovarian cancer cells. Two reporter constructs driven by either the entire (P1+P2) promoter or only the P2 promoter of bcl-2 gene (Fig. 2A) were used to determine their promoter activity in the presence of absence of DDB2 overexpression in CP70 cells, which express negligible endogenous DDB2 (31). As shown in Fig. 2B, DDB2 transient overexpression reduced the bcl-2 promoter activity by nearly 30% when the entire bcl-2 promoter region, but not the P2 region only, was present in the reporter assay system. This indicates that DDB2 is able to down-regulate bcl-2 promoter activity to impose its transcriptional repression on bcl-2.
Figure 2.
DDB2 inhibits the P1 promoter activity of bcl-2 gene. A, the schematic diagram of bcl-2 gene promoter structure. TSS: Translation start site. B, two luciferase reporters containing either P1+P2 or P2 only, empty or DDB2-expressing vector were transfected into CP70 cells. The relative promoter activity was calculated by normalizing to the empty vector (set as 100%). C, CP70 cells were transfected with DDB2-WT, DDB2-K244E, or empty vector for 48 h. The relative mRNA level was quantified by real-time PCR using GAPDH as endogenous control, and normalized to the empty vector (set as 100%). The upper panel represents DDB2 expression level in the transfected cells by immunoblotting using Tubulin as the loading control. D, CP70 cells were transfected with reporter construct P1+P2, DDB2-WT, DDB2-K244E, or empty vector for 48 h, the promoter activity was analyzed as in (B). n=3, bar: SD, *: p<0.05 by 2-sample t-test.
DDB2 is known to bind to the damaged DNA in a sequence-independent manner (24, 25). One natural occurring mutant DDB2-K244E, identified from XP-E patients, is known to be defective in DNA-binding (36, 37). To understand whether the DNA binding capacity of DDB2 is required for the transcriptional repression of bcl-2, a DDB2-K244E expression vector was used in the reporter assay. As shown in Fig. 2C, the DDB2-K244E mutant failed to suppress bcl-2 transcription. Consistent with this finding, the bcl-2 promoter activity was not affected by overexpression of DDB2-K244E mutant (Fig. 2D). Therefore, it appears that DNA binding capacity of DDB2, primarily through lysine 244 located in the β-propeller domain of DDB2, plays an essential role in DDB2-mediated bcl-2 transcriptional regulation.
A putative DDB2 binding site was identified in the bcl-2 P1 promoter
Having established that bcl-2 is subject to DDB2-mediated transcriptional modulation through the P1 promoter, we sought to identify cis-acting elements that are specifically recognized and bound by DDB2 in this region. DDB2 was recently revealed to repress MnSOD2 transcription via a cis-response element 5′-AGCCTGCAGCCT-3′ located in the proximal promoter of the MnSOD2 gene (38). Aided by this information, we performed a thorough sequence alignment with the above DDB2 binding sequence across the entire bcl-2 P1 promoter region, as well as the boundary region between these two promoters using DNASTAR software (DNASTAR, Inc., Madison, WI). Two putative DDB2 binding sites were identified: one is located at the extreme 5′ end of bcl-2 P1 promoter, which differs by only 2 nucleotides with the reported sequence (labeled as BCP1 in Fig. 3A), and the second lies in the boundary between P1 and M region with the sequence half split in each section (labeled as BCPM in Fig. 3A). Quantitative ChIP analysis was then used to investigate whether DDB2 binds to these sites in vivo. As shown in Fig. 3B, DDB2 was enriched by 2.7-fold in the BCP1 site, but not in BCPM and two other sites (−2622 to −2484 and −2750 to −2611 in the bcl-2 promoter, data not shown).
Figure 3.
DDB2 binds directly to a specific cis-acting response element in bcl-2 P1 promoter. A, the schematic diagram of bcl-2 gene promoter structure and sequence alignment between two putative DDB2 binding sites in bcl-2 gene and the DDB2-binding site in MnSOD gene. The two putative binding sites are named as BCP1 (−3804 to −3814 in bcl-2 promoter) and BCPM (−1346 to −1355 in bcl-2 promoter). B, recruitment of DDB2 to BCP1 and BCPM sites in bcl-2 promoter by quantitative ChIP analysis in HeLa and HeLa-DDB2 cells. The relative fold enrichment was quantified by normalization to input first, then normalized to HeLa cells, which is set at 1. n=3, bar: SD, *: p < 0.01. C, D, E, DDB2 binds to the BCP1 site in vitro. (C) DDB.com was purified from HeLa-DDB2 cells as described in the “Material and Methods”. The purified complexes were resolved by SDS-PAGE and visualized by Coomassie blue staining. (D) EMSA assay: 50 fmol IRDye 700 5′-end-labeled DNA probe (either the Bcl-2 probe containing the BCP1 site or the MnSOD probe containing the reported DDB2 binding site), 1 μg poly(dI-dC), 0.9 or 1.5 μg DDB.com were incubated for 35 min at 25°C in the dark for the EMSA assay. (E) Super-shift EMSA assay: 1 μg poly(dI-dC), 1.4 μg DDB.com, 1 μl goat anti-DDB2 antibody were incubated for 15 min on ice in the dark before another 15 min incubation with 50 fmol Bcl-2 probe at 25°C in the dark for the EMSA assay. (F) Cold EMSA competition assay: 50 fmol Bcl-2 probe, 0.5 μg poly(dI-dC), 2 μg DDB.com, 10 pmol of of unlabeled oligos containing the wild-type BCP1 site (Ow) or mutant BCP1 site (Om) were incubated in a 20 μl reaction for 1 h at 37°C in the dark for the EMSA assay. The forward sequences of Ow and Om oligos were shown in the left panel where the BCP1 site was underlined and the mutations are shown in italic form (D). NS: non-specific band. The protein-DNA complex was indicated by an arrow.
To further confirm BCP1 as a bona fide DDB2 response element, EMSA was performed with a 24-bp oligo corresponding to the region from −3797 to −3820 in bcl-2 P1 promoter flanking the BCP1 site, and the DDB2 protein in complex with DDB1-Cul4A-Roc1 (DDB2.com), purified from HeLa-DDB2 cells (Fig. 3C). As seen in Fig. 3D, a slower-migrating species appeared following the addition of DDB.com (lane 3). Similarly, EMSA with the MnSOD probe and DDB.com also showed a slower-migrating band (lane 6) as reported by Minig V. et al (38), verifying our EMSA assay and the activity of DDB.com. In addition, the slower-migrating band disappeared in the super-shift assay after anti-DDB2 antibody was added to the system (Fig. 3E), indicating it is the DDB2 subunit in the DDB2.com that specifically binds to the oligo. This observation is also in line with the report that only DDB2 harbors the intrinsic DNA-binding capability among all DDB2-containing forms identified to date (39). To verify the sequence specificity of DDB2-DNA binding, two unlabeled specific competitors were used to compete with the target probe in EMSA: one is of identical sequence with the probe (Ow), and the other (Om) carries a GC to TA transversion in the first AGCCT repeat of the BCP1 site. This sequence was chosen because it is reported that the sequence in the two pentanucleotide repeats is critical for DDB2 binding (38). As expected, addition of Ow results in the disappearance of the DDB.com-DNA complex, but no effect was observed when Om was used for competition (Fig. 3F). In summary, our data indicates that DDB2 binds to the BCP1 site in the bcl-2 promoter in a sequence-specific manner both in vivo and in vitro.
Overexpression of DDB2 promotes histone H3 deacetylation in the bcl-2 promoter and enhancer regions
We then asked how the binding of DDB2 to the bcl-2 promoter affects the local chromatin structure to inhibit bcl-2 transcription. One possible mechanism is through spatial interference of the accessibility of other transcription factors. However, quantitative ChIP analysis demonstrated that neither E2F1 nor Sp1 exhibited any significant change in local enrichment to the bcl-2 P1 core promoter when DDB2 is overexpressed (Fig. S2A, B), suggests that DDB2 acts independently of both E2F1 and Sp1 to negatively control the bcl-2 transcription.
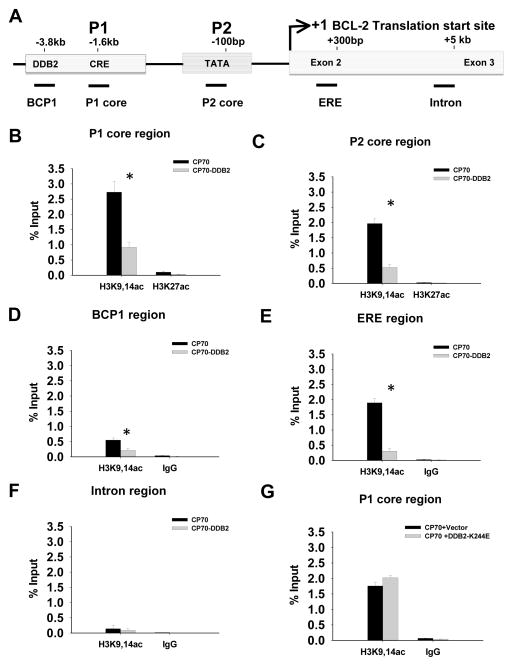
Then we turned our focus to histone modification status along the regulatory regions in the bcl-2 gene. Five specific regions in bcl-2 (Fig. 4A) were selected for quantitative ChIP analyses in CP70 and CP70-DDB2 cells: BCP1 region (flanking the BCP1 site), P1 core (flanking CRE and Sp1 sites), P2 core (flanking the TATA and CAAT box), ERE (flanking the two Estrogen-responsive elements), and an intron region 5 kb downstream of the bcl-2 transcriptional start site. A significant decrease in the acetylation level of histone H3K9, 14 was detected in all of the selected regions except for the intron region when DDB2 was overexpressed (Fig. 4B–F). In contrast, overexpression of mutated DDB2 (K224E), which is unable to bind to DNA, failed to reduce the enrichment of H3K9,14ac in the bcl-2 P1 promoter (Fig. 4G). Interestingly, in both P1 and P2 core promoters, no significant difference in H3K27 acetylation level was identified between CP70 and CP70-DDB2 cells (Fig. 4B–C). Taken together, this data indicates that the elevated histone H3 deacetylation induced by DDB2 overexpression takes place across the regulatory region of bcl-2 in a sequence- and lysine site-specific manner.
Figure 4.
DDB2 affects histone H3 acetylation status at bcl-2 promoter and enhancer regions. A, the schematic diagram of bcl-2 gene structure. The selected regions for ChIP analysis were marked by thick lines underneath the diagram with names labeled below correspondingly. B–F, CP70 and CP70-DDB2 cells were used to compare the local enrichment of H3K9, 14ac and H3K27ac in specific regions as indicated in (A). The fraction bound by specific antibodies was calculated by normalization to the input. n=3, bar: SD, *: p < 0.01. G, CP70 cells were transiently transfected with empty vector or DDB2-K244E for 72 h, ChIP assay was conducted to detect the enrichment of H3K9, 14ac in the bcl-2 P1 core region.
HDAC1 is recruited to the bcl-2 P1 promoter by DDB complex through direct interaction with DDB1
Alterations in the activities either of histone acetyltransferase (HAT) and/or histone deacetylases (HDAC) can affect histone acetylation levels. The recruitment of histone deacetylase to the bcl-2 promoter (40) have been reported. Thus, the enrichment profile of HDAC1 and HDAC2 in the bcl-2 P1 and P2 core promoter regions was mapped out in CP70 and CP70-DDB2 cells by quantitative ChIP analyses. No significant difference in the local enrichment of HDAC2 to the bcl-2 promoter was detected between CP70 and CP70-DDB2 cells. In contrast, HDAC1 exhibited a significant enrichment in the P1, but not P2 core promoter region in the cells overexpressing DDB2 (Fig. 5A, B). Conversely, DDB2 knock-down by lentiviral-mediated RNA interference in A2780 cells (cisplatin-sensitive, DDB2-proficient cells) reduced HDAC1 accumulation in the P1 core promoter region of bcl-2 gene (Fig. 5D, E). This prompted us to hypothesize that HDAC1 might be recruited by DDB complex to the bcl-2 P1 promoter region. To test this hypothesis, we investigated a possible in vivo interaction between HDAC1 and DDB complex. We pulled down the intact DDB complex with anti-FLAG antibody in the cells transiently or stably overexpressing FLAG-tagged DDB2. In both systems, HDAC1 was found to reside in the DDB complex (Fig. 6A). However, this in vivo assay is unable to address the question whether DDB2 or DDB1 directly interacts with HDAC1. To answer this question, we proceed with an in vitro binding assay using purified recombinant proteins to examine a potential direct interaction between HDAC1 and DDB1 or DDB2. FLAG-tagged DDB1 and FLAG-tagged DDB2 were purified from insect Sf9 cells by one-step FLAG affinity purification (Fig. S3), and incubated with bacterially expressed, GST affinity -purified GST or GST-HDAC1 (Fig. S3). As shown in Fig. 6B, FLAG-DDB1 was specifically pulled down by GST-HDAC1 but not GST in the binding assay. In contrast, GST-HDAC1 failed to pull down FLAG-DDB2. This data indicates a specific and direct interaction between HDAC1 and DDB1 in vitro.
Figure 5.
DDB2 recruits HDAC1 to bcl-2 P1 core promoter region. A, B, ChIP assay was conducted in CP70 and CP70-DDB2 cells to estimate the enrichment of HDAC1 and HDAC2 on P1 (A) and P2 (B) core promoter region. The relative fold enrichment was calculated by normalization to the input first, and then normalized to the negative control IgG (set as 1). n=3, bar: SD, *: p < 0.05. C, D, A2780 cells were transduced with shCtrl or shDDB2 lentiviruses at MOI=2.5 for 48 h, the expression of DDB2 was detected with anti-DDB2 antibody. Lamin B was used as a loading control (C). ChIP assay was conducted to estimate the enrichment of HDAC1 on bcl-2 P1 core promoter region. n=3, bar: SD, *: p < 0.05.
Figure 6.
DDB1 interacts with HDAC1 directly. A, DDB complex was immunoprecipitated with anti-FLAG affinity gel from CP70 cells transfected with FLAG-DDB2 or mock-transfected (left panel), and HeLa or HeLa-DDB2 cells (right panel), the presence of DDB1, DDB2 and HDAC1 in the immunoprecipitates was detected using immunoblotting with anti-DDB1, anti-DDB2 and anti-HDAC1 antibodies, respectively. Arrow: IgG heavy chain. B, GST binding assay was conducted to determine the direct interaction between HDAC1 and DDB1 or DDB2. C, D, CP70 cells was transduced with shCtrl or shHDAC1 lentivirus at MOI =2.5 for 48 h, the expression of HDAC1 was detected using immunoblotting with anti-HDAC1 antibody. Ponceau S staining serves as the loading control (C). The luciferase reporter containing the entire bcl-2 promoter was transfected into shCtrl and shHDAC1 transduced CP70 cells. The relative promoter activity was calculated by normalizing to the shCtrl-transduced cells (set as 100%). n=3, bar: SD, **: p<0.01 (D).
To further validate the role of HDAC1 in DDB complex-mediated bcl-2 transcriptional down-regulation, we compared the bcl-2 promoter activity in the cells with or without HDAC1 knockdown. As detected by the dual luciferase reporter assay (Fig. 6C, D), the bcl-2 promoter activity was enhanced by nearly 1.3 fold following HDAC1 knockdown in comparison with the control, confirming that HDAC1 is involved in DDB complex-mediated bcl-2 transcriptional inhibition.
DDB2-mediated repression of bcl-2 expression enhances cisplatin-induced apoptosis
To understand the biological significance of DDB2-mediated repression of Bcl-2 transcription, we overexpressed either DDB2-WT or DDB2-K244E in CP70 cells and assessed the cellular apoptosis upon cisplatin treatment. As shown in Fig. 7, DDB2-WT enhanced cisplatin-induced apoptosis as reflected by the increased cleaved PARP and cleaved caspase-3 (Fig. 7A, B) as well as increased Annexin V-positive cells (Fig. 7C, D). However, cisplatin-induced apoptosis is compromised in DDB2-K244E overexpressing CP70 cells compared with DDB2-WT overexpressing cells (Fig. 7). Consistently, Bcl-2 expression is dramatically reduced in DDB2-WT expressed cells, while is only slightly reduced in DDB2-K244E expressed cells (Fig. 7A, B). Given that DDB2-K244E is unable to bind to the bcl-2 promoter to repress Bcl-2 transcription, the difference in cisplatin-induced apoptosis between DDB2-WT and DDB2-K244E overexpressed cells can be attributed to DDB2-mediated alteration of Bcl-2 expression.
Figure 7.
DDB2-mediated downregulation of Bcl-2 enhanced cisplatin-induced apoptosis in CP70 cells. A, B, CP70 cells were transfected with either DDB2-WT, DDB2-K244E, or empty vector for 24 h, then treated with 60 μM cisplatin for 1 h and further cultured in drug-free medium for another 48 h. Whole cell lysates were prepared and subjected to immunoblotting for the detection of DDB2, Bcl-2, cleaved PARP, and cleaved Caspase-3. A, representative Western blots of three independent experiments. B, quantification of the band intensity of cleaved PARP, cleaved Caspase-3 and Bcl-2 expression at 48 h after cisplatin treatment by using ImageJ, n =3, Bar: SD, *: p<0.01. C, D, transfected CP70 cells were treated with 60 or 120 μM cisplatin for 1 h and further cultured in drug-free medium for another 24 h. Apoptotic cells were detected with Annexin V staining. C, representative flow cytometry analysis plots. D, the average percentage of Annexin V-positive cells from 3 independent repeats. Bar: SD, *: p<0.01
Discussion
Bcl-2, a key regulator in apoptosis and carcinogenesis, is frequently overexpressed in cisplatin-resistant ovarian cancer cells (2, 41). The bcl-2 regulation network is complex and not fully understood to date. We reported previously that DDB2 exhibits a negative correlation with Bcl-2 at both the transcriptional and translational level in several cisplatin-resistant ovarian cancer cell lines (31). Here we discussed the mechanism through which DDB1 and DDB2 subunits in the DDB complex cooperate to negatively regulate bcl-2 transcription in ovarian cancer cells.
Two promoters, P1 and P2, have been identified for the bcl-2 transcription regulation. In the majority of cell types, bcl-2 transcription is regulated through the P1 promoter (3, 5, 6, 8). This appears to be the case in ovarian cancer cells wherein DDB2 recognizes a responsive element located at the extreme 5′ end of bcl-2 P1 promoter to exert the repressive effect. In light of the observation that K244E mutant of DDB2 fails to affect bcl-2 transcription, we propose that lysine 244 is indispensable for mediating the association between DDB2 and the bcl-2 promoter, which is further supported by the ChIP analysis showing that K244E mutant of DDB2 had no effect on the local histone H3K9, 14 deacetylation at bcl-2 promoter. As revealed by structural analyses, K244 residue in DDB2 is in direct contact with the DNA phosphodiester backbone via charge-stabilized hydrogen bonds (24), thus it is not surprising that this single residue in DDB2 is important for two seemingly conflicting tasks: the canonical sequence-independent binding to damaged DNA in GGR and the sequence-specific binding to the BCP1 site in the bcl-2 promoter.
Direct binding of DDB2 to the BCP1 site evidenced in vivo and in vitro by the ChIP and EMSA assay suggests that DDB2 functions as a transcriptional repression factor for bcl-2. It has been previously reported that DDB2 is a negative transcription factor for MnSOD (38) (42), catalase (42), VEGF, Zeb1 and Snail (43). The DDB2 binding site in bcl-2 gene differs only by 2 nucleotides from that in the MnSOD gene. Considering that MnSOD is important for cell growth in breast cancer cells while Bcl-2 is a survival factor in multiple cancers types including ovarian cancer, the binding of DDB2 to such a consensus site may imply a more general theme of DDB2 to exert transcriptional control over genes important in cell proliferation. DDB2 binding is coupled with histone H3 deacetylation in all cases; however, H3K27 was targeted for deacetylation in MnSOD (38), while H3K9,14, not H3K27, were specially selected for deacetylation in the bcl-2 promoter.
By mapping the enrichment profile of two HDAC proteins to the bcl-2 promoter region, HDAC1 seems to be an important candidate responsible for the observed H3 de-acetylation activities along the bcl-2 regulatory regions. It might be of note that the widespread deacetylation effect observed across the bcl-2 promoter region, not limiting to the P1 core promoter region where HDAC1 is recruited, may be partially explained by the spatial proximity between HDAC1 to its targets brought about by its recruitment. It is also possible that other deacetylases than HDAC1 might also contribute to this effect.
Knockdown of HDAC1 leads to an increased promoter activity of bcl-2 in CP70 cells overexpressing DDB2, arguing strongly for the importance of HDAC1 in DDB complex mediated bcl-2 transcriptional regulation. Duan H et al have reported that HDAC inhibitors downregulate bcl-2 expression in t(14;18) lymphoma cells DHL-41 through inhibiting bcl-2 promoters (40). It looks like this finding and ours demonstrate opposite effect of HDACs on bcl-2 promoter. However, Duan et al also demonstrated that even though HDAC inhibitors increased overall acetylation of histones, localized acetylated histone H3 decreased and HDAC1 recruitment increased at both bcl-2 promoters (40). Therefore, the recruitment of HDAC1 to the bcl-2 promoters seems a common mechanism for the repression of Bcl-2 expression.
Both subunits of the DDB complex contribute to the HDAC1 recruitment to the bcl-2 promoter region in ovarian cancer cells. The importance of DDB2 in this regard might largely arise from its role in anchoring the downstream factors in place. Consequently, DDB2 overexpression and knockdown significantly affected HDAC1 local recruitment. The contribution of DDB1 appears to come from its direct interaction with HDAC1, as shown by the in-vitro binding assay. Based on our current understanding of the DDB-mediated bcl-2 transcriptional regulation mechanism in ovarian cancer cells, we propose a model whereby DDB2 in the DDB complex is responsible for recognizing and binding to the BCP1 site in the 5′-end of the bcl-2 P1 promoter, HDAC1 was then recruited, most likely through DDB1, to the P1 core region located 2.2 kb downstream of BCP1 and deacetylates histone H3 at lysine 9, 14 sites along the regulatory regions of bcl-2, resulting in bcl-2 transcriptional repression.
We have demonstrated a stronger negative correlation between DDB2 and Bcl-2 protein levels than that between DDB2 protein and Bcl-2 mRNA levels in human tumor samples. It seems that DDB2 may also affect Bcl-2 protein stability. Indeed, we have previously shown that proteasome degradation is involved in the DDB2-mediated Bcl-2 reduction, probably through DDB-Cul4A E3 ligase-mediated ubiquitylation (31). In addition, although abolishing DNA binding capacity of DDB2 compromises DDB2-mediated apoptosis upon cisplatin treatment, DDB2-K244E mutant still slightly enhances cisplatin-induced apoptosis, indicating an alternative Bcl-2-independent mechanism exists, and warranting a further investigation.
Supplementary Material
Acknowledgments
Financial support: This work was supported by the National Institute of Health grant CA151248 to Q.E. Wang and CA93413 to A.A. Wani and Q.E. Wang.
The authors thank Drs. Yoshihiro Nakatani (Harvard University) and Paul Modrich (Duke University) for kindly providing cells, Drs. Gilbert Chu (Stanford University), Yi Qiu (University of Florida) and Linda Boxer (Stanford University) for providing plasmids, Dr. Richard D. Wood (M.D. Anderson Cancer Center) for providing baculoviruses, Drs. David Cohn and Brent Tierney (The Ohio State University) for providing tumor tissues. Dr. Ying Wang (The Ohio State University) for providing valuable suggestions for EMSA.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed
Authors’ Contributions
Conception and design: R.Zhao, Q.E. Wang
Development of methodology: R.Zhao, C.Han, Q.E.Wang
Acquisition of data (provided animals acquired and managed patients, provided facilities, etc.): R.Zhao, C.Han, E.Eisenhauer, Q.E.Wang
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): R.Zhao, Q.E.Wang
Writing, review, and/or revision of the manuscript: R.Zhao, E.Eisenhauer, A.A.Wani, Q.E.Wang
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.Kroger, W.Zhao, K.Selvendiran, J.Yu, X.Liu
Study supervision: Q.E.Wang
References
- 1.Ozols RF. Ovarian cancer: new clinical approaches. Cancer Treat Rev. 1991;18(Supplement A0):77–83. doi: 10.1016/0305-7372(91)90027-w. [DOI] [PubMed] [Google Scholar]
- 2.Basu A, Haldar S. The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Mol Hum Reprod. 1998;4(12):1099–109. doi: 10.1093/molehr/4.12.1099. [DOI] [PubMed] [Google Scholar]
- 3.Huang H, Zegarra-Moro OL, Benson D, Tindall DJ. Androgens repress Bcl-2 expression via activation of the retinoblastoma (RB) protein in prostate cancer cells. Oncogene. 2004;23(12):2161–76. doi: 10.1038/sj.onc.1207326. [DOI] [PubMed] [Google Scholar]
- 4.Yunis J. The chromosomal basis of human neoplasia. Science. 1983;221(4607):227–36. doi: 10.1126/science.6336310. [DOI] [PubMed] [Google Scholar]
- 5.Young RL, Korsmeyer SJ. A negative regulatory element in the bcl-2 5′-untranslated region inhibits expression from an upstream promoter. Mol Cell Biol. 1993;13(6):3686–97. doi: 10.1128/mcb.13.6.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16(10):5546–56. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Q, Li X, Vale-Cruz D, Brown ML, Beier F, LuValle P. Activating transcription factor 2 controls Bcl-2 promoter activity in growth plate chondrocytes. J Cell Biochem. 2007;101(2):477–87. doi: 10.1002/jcb.21198. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Manzano C, Mitlianga P, Fueyo J, Lee H-Y, Hu M, Spurgers KB, et al. Transfer of E2F-1 to Human Glioma Cells Results in Transcriptional Up-Regulation of Bcl-2. Cancer Res. 2001;61(18):6693–7. [PubMed] [Google Scholar]
- 9.Heckman C, Mehew J, Boxer L. NF-kappaB activates Bcl-2 expression in t(14;18) lymphoma cells. Oncogene. 2002;21(24):3898–908. doi: 10.1038/sj.onc.1205483. [DOI] [PubMed] [Google Scholar]
- 10.Heckman CA, Mehew JW, Ying G-G, Introna M, Golay J, Boxer LM. A-Myb Up-regulates Bcl-2 through a Cdx Binding Site in t(14;18) Lymphoma Cells. J Biol Chem. 2000;275(9):6499–508. doi: 10.1074/jbc.275.9.6499. [DOI] [PubMed] [Google Scholar]
- 11.Heckman CA, Wheeler MA, Boxer LM. Regulation of Bcl-2 expression by C//EBP in t(14;18) lymphoma cells. Oncogene. 2003;22(39):7891–9. doi: 10.1038/sj.onc.1206639. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y-l, Mehew JW, Heckman CA, Arcinas M, Boxer LM. Negative regulation of bcl-2 expression by p53 in hematopoietic cells. Oncogene. 2001;20:240–51. doi: 10.1038/sj.onc.1204067. [DOI] [PubMed] [Google Scholar]
- 13.Svotelis A, Bianco S, Madore J, Huppe G, Nordell-Markovits A, Mes-Masson A-M, et al. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ER[alpha] ligand dependency. EMBO J. 2011;30(19):3947–61. doi: 10.1038/emboj.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong F, Sun L, Wang Z, Shi J, Li W, Wang S, et al. The BCL2 gene is regulated by a special AT-rich sequence binding protein 1-mediated long range chromosomal interaction between the promoter and the distal element located within the 3′-UTR. Nucleic Acids Research. 2011;39(11):4640–52. doi: 10.1093/nar/gkr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dualan R, Brody T, Keeney S, Nichols AF, Admon A, Linn S. Chromosomal Localization and cDNA Cloning of the Genes (DDB1 and DDB2) for the p127 and p48 Subunits of a Human Damage-Specific DNA Binding Protein. Genomics. 1995;29(1):62–9. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 16.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242(4878):564–7. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 17.Hwang BJ, Toering S, Francke U, Chu G. p48 Activates a UV-Damaged-DNA Binding Factor and Is Defective in Xeroderma Pigmentosum Group E Cells That Lack Binding Activity. Mol Cell Biol. 1998;18(7):4391–9. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh T, Linn S, Ono T, Yamaizumi M. Reinvestigation of the classification of five cell strains of xeroderma pigmentosum group E with reclassification of three of them. The Journal Of Investigative Dermatology. 2000;114(5):1022–9. doi: 10.1046/j.1523-1747.2000.00952.x. [DOI] [PubMed] [Google Scholar]
- 19.Rapić-Otrin V, Navazza V, Nardo T, Botta E, McLenigan M, Bisi DC, et al. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum Mol Genet. 2003;12(13):1507–22. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- 20.Tang J, Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair. 2002;1(8):601–16. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittschieben BØ, Iwai S, Wood RD. DDB1-DDB2 (Xeroderma Pigmentosum Group E) Protein Complex Recognizes a Cyclobutane Pyrimidine Dimer, Mismatches, Apurinic/Apyrimidinic Sites, and Compound Lesions in DNA. J Biol Chem. 2005;280(48):39982–9. doi: 10.1074/jbc.M507854200. [DOI] [PubMed] [Google Scholar]
- 22.Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H. Regulation of Nucleotide Excision Repair by UV-DDB: Prioritization of Damage Recognition to Internucleosomal DNA. PLoS Biol. 2011;9(10):e1001183. doi: 10.1371/journal.pbio.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groisman R, Polanowska J, Kuraoka I, Sawada J-i, Saijo M, Drapkin R, et al. The Ubiquitin Ligase Activity in the DDB2 and CSA Complexes Is Differentially Regulated by the COP9 Signalosome in Response to DNA Damage. Cell. 2003;113(3):357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 24.Scrima A, Koní ková R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, et al. Structural Basis of UV DNA-Damage Recognition by the DDB1 DDB2 Complex. Cell. 2008;135(7):1213–23. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer Eric S, Scrima A, Böhm K, Matsumoto S, Lingaraju Gondichatnahalli M, Faty M, et al. The Molecular Basis of CRL4DDB2/CSA Ubiquitin Ligase Architecture, Targeting, and Activation. Cell. 2011;147(5):1024–39. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 26.Hayes S, Shiyanov P, Chen X, Raychaudhuri P. DDB, a Putative DNA Repair Protein, Can Function as a Transcriptional Partner of E2F1. Mol Cell Biol. 1998;18(1):240–9. doi: 10.1128/mcb.18.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, et al. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutation Research/DNA Repair. 2001;486(2):89–97. doi: 10.1016/s0921-8777(01)00082-9. [DOI] [PubMed] [Google Scholar]
- 28.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, et al. Human STAGA Complex Is a Chromatin-Acetylating Transcription Coactivator That Interacts with Pre-mRNA Splicing and DNA Damage-Binding Factors In Vivo. Mol Cell Biol. 2001;21(20):6782–95. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc Natl Acad Sci. 2009;106(26):10690–5. doi: 10.1073/pnas.0812254106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoyanova T, Yoon T, Kopanja D, Mokyr MB, Raychaudhuri P. The Xeroderma Pigmentosum Group E Gene Product DDB2 Activates Nucleotide Excision Repair by Regulating the Level of p21Waf1/Cip1. Mol Cell Biol. 2008;28(1):177–87. doi: 10.1128/MCB.00880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barakat BM, Wang Q-E, Han C, Milum K, Yin D-T, Zhao Q, et al. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int J Cancer. 2010;127(4):977–88. doi: 10.1002/ijc.25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q-E, Prætorius-Ibba M, Zhu Q, El-Mahdy MA, Wani G, Zhao Q, et al. Ubiquitylation-independent degradation of Xeroderma pigmentosum group C protein is required for efficient nucleotide excision repair. Nucleic Acids Res. 2007;35(16):5338–50. doi: 10.1093/nar/gkm550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Q, Wang Q-E, Ray A, Wani G, Han C, Milum K, et al. Modulation of Nucleotide Excision Repair by Mammalian SWI/SNF Chromatin-remodeling Complex. J Biol Chem. 2009;284(44):30424–32. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen TN, Goodrich JA. Protein-protein interaction assays: eliminating false positive interactions. Nat Methods. 2006;3(2):135. doi: 10.1038/nmeth0206-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hummon A, Lim S, Difilippantonio M, Ried T. Isolation and solubilization of proteins after TRIzol® extraction of RNA and DNA from patient material following prolonged storage. Biotechniques. 2007;42(4):467–72. doi: 10.2144/000112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols AF, Ong P, Linn S. Mutations Specific to the Xeroderma Pigmentosum Group E Ddb Phenotype. J Biol Chem. 1996;271(40):24317–20. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda N, Azuma K, Saijo M, Iemura S-i, Hioki Y, Natsume T, et al. DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by Cullin 4A-based ubiquitin ligase complex. DNA Repair. 2005;4(5):537–45. doi: 10.1016/j.dnarep.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Minig V, Kattan Z, van Beeumen J, Brunner E, Becuwe P. Identification of DDB2 Protein as a Transcriptional Regulator of Constitutive SOD2 Gene Expression in Human Breast Cancer Cells. J Biol Chem. 2009;284(21):14165–76. doi: 10.1074/jbc.M808208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulaksiz G, Reardon JT, Sancar A. Xeroderma Pigmentosum Complementation Group E Protein (XPE/DDB2): Purification of Various Complexes of XPE and Analyses of Their Damaged DNA Binding and Putative DNA Repair Properties. Mol Cell Biol. 2005;25(22):9784–92. doi: 10.1128/MCB.25.22.9784-9792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan H, Heckman CA, Boxer LM. Histone Deacetylase Inhibitors Down-Regulate bcl-2 Expression and Induce Apoptosis in t(14;18) Lymphomas. Mol Cell Biol. 2005;25(5):1608–19. doi: 10.1128/MCB.25.5.1608-1619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Zhou J-Y, Zhang L, Wu GS. Involvement of MKP-1 and Bcl-2 in acquired cisplatin resistance in ovarian cancer cells. Cell Cycle. 2009;8(19):3191–8. doi: 10.4161/cc.8.19.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy N, Stoyanova T, Dominguez-Brauer C, Park HJ, Bagchi S, Raychaudhuri P. DDB2, an Essential Mediator of Premature Senescence. Mol Cell Biol. 2010;30(11):2681–92. doi: 10.1128/MCB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Roy N, Bommi PV, Bhat UG, Bhattacharjee S, Elangovan I, Li J, et al. DDB2 Suppresses Epithelial to Mesenchymal Transition in Colon Cancer. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.