Table 1.
Experimental affinities and free energy calculation blind predictions for compounds tested in the CCP open cavity.
| Round 1 Compound | Exper. ΔGbind (kcal/mol) |
Predicted ΔGbind (kcal/mol) |
Predicted pose RMSD (Å) |
PDB Code | |
|---|---|---|---|---|---|
| 1 | 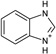 |
−5.8a | −5.8 ± 0.1b | 1.1c | 1KXM |
| 2 | 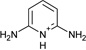 |
−5.8 ± 0.2 | −5.1 ± 0.2 | 0.6 | 4JM8 |
| 3 |  |
−5.1 ± 0.2 | −4.8 ± 0.2 | 1.9 | 4JM5 |
| 4 | −4.4 ± 0.2 | −2.2 ± 0.2 | 3.1 | 4JM6 | |
| 5 | 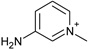 |
−3.4 ± 0.4 | −1.1 ± 0.2 | 2.9 (1st) 0.5 (2nd) |
4JM9 |
| Round 2 Compound | Exper. ΔGbind (kcal/mol) |
Predicted ΔGbind (kcal/mol) |
Predicted pose RMSD (Å) |
PDB Code | |
| 6 |  |
−7.1 ± 0.2 | −4.2 ± 0.3 | 0.6 | 4JQM |
| 7 | 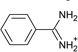 |
−6.6 ± 0.2 | −3.3 ± 0.2 | 0.5 | 4JPU |
| 8 | 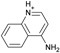 |
−5.8 ± 0.2 | −5.8 ± 0.3 | 0.4 | 4JQJ |
| 9 | 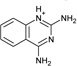 |
−5.7 ± 0.2 | −4.7 ± 0.4 | 0.9 | 4JPT |
| 10 | −4.8 ± 0.2 | −7.9 ± 0.4 | 1.0 | 4JPL | |
| (Corrected Tyr) | −4.8 ± 0.2 | −5.9 ± 0.4d | 3.2e | 4JPL | |
| 11 |  |
−4.7 ± 0.2 | −2.3 ± 0.3 | 1.1 | 4JQK |
| 12 |  |
> −3.9 | −3.7 ± 0.4 | naf | na |
| 13 | 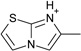 |
> −3.9 | −2.8 ± 0.2 | na | na |
| 14 | 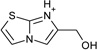 |
> −3.0 | −4.4 ± 0.5 | na | na |
| 15 | NB, > 3.0 | −0.9 ± 0.5 | na | na | |
| Neutral Compound | Exper. ΔGbind (kcal/mol) |
Predicted ΔGbind (kcal/mol) |
Predicted pose RMSD (Å) |
PDB Code | |
| 16 | > −3.3 | −4.6 ± 0.3 | 1.0 | 4JMW | |
| 17 | > −3.3 | −4.8 ± 0.2 | 2.8 | 4JMA | |
| 18 | > −3.3 | −2.6 ± 0.4 | 2.8 (1st) 0.8 (2nd) |
4JQN | |
| 19 | NB, > −3.9 | −6.8 ± 0.3 | na | na | |
| 20 | NB, > −3.0 | −5.2 ± 0.3 | na | na | |
NB = no evidence of binding, to a given maximum concentration. Round One predictions were made using a scaling factor of +/− 0.981, calibrated using benzimidazole (compound 1). Round Two predictions were made using a scaling factor of 0.986, calibrated based on the five Round One compounds. Pose RMSDs were calculated using a representative snapshot of the MD ensemble for the dominant pose, measured against the nearest ligand structure observed by crystallography after aligning the MD protein to the crystal protein. Experimental uncertainties were calculated as the standard deviation of three or more independent measurements, or as a 20% error in Kd if only two experiments were conducted or if the experimental agreement was within 20%.
From ref. 7.
Not a blind prediction, used to calibrate the original scaling factor.
Not a blind prediction, but the RMSD to the most favored benzimidazole pose of several decoys considered.
Not a blind prediction; based on re-simulating the badly equilibrated 4-azaindole pose retrospectively.
Not a blind prediction, but based on using the most favorable 4-azaindole pose after retrospectively resimulating the badly equilibrated 4-azaindole pose.
Not applicable (no structure collected).
