Figure 6.
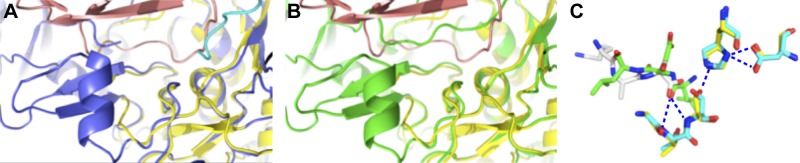
Comparison of active sites of rEpiP and rEpiP-S393A proteins with a thermitase-eglin-c complex. A) Overlay of rEpiP and a thermitase-eglin-c complex (PDB ID: 2TEC). B) Overlay of the rEpiP-S393A and thermitase-eglin-c complex. rEpiP is shown in blue, rEpiP-S393A in green, and the thermitase-eglin-c complex in pink. The C terminus of a neighboring molecule in the rEpiP active site is shown in cyan. C) Ball-and-stick representation of the superposition of the active sites of the thermitase-eglin-c complex and rEpiP. Nitrogen atoms are shown in blue, oxygen atoms in red, carbon atoms for the thermitase in yellow and for rEpiP (chain A) in cyan, the thermitase-eglin-c complex in green, and the C-terminal portion of chain H of rEpiP in gray. Despite the fact that peptide chains for thermitase-eglin-c complex and rEpiP chain H run in the opposite direction, carbonyl oxygen atoms are pointed in the same direction. The position of one of the C-terminal oxygens of rEpiP chain H matches the position of the carbonyl oxygen of the thermitase-eglin-c complex, which is bound in the oxyanion hole. There are almost no differences in the active sites of rEpiP and rEpiP-S393A, so the mutant is not shown here. In the crystal structure of rEpiP, the active site of the protein contains the C terminus of a neighboring molecule as follows: in the active site of A–H is the C terminus of chains H, D, F, G, B, A, E, and C, respectively.
