Figure 4.
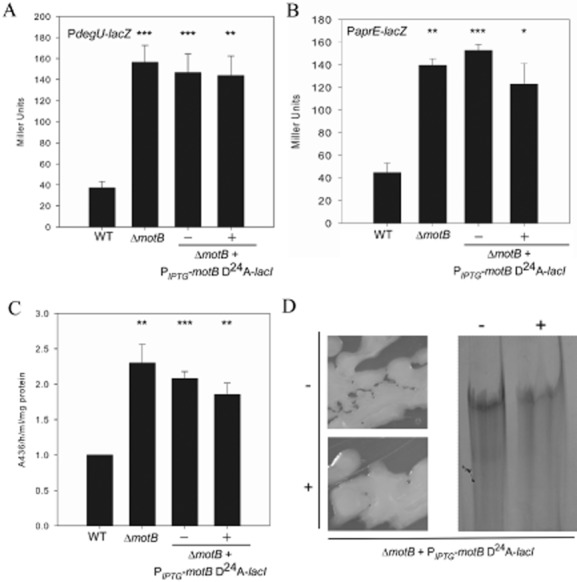
The motB D24A allele cannot complement ΔmotB.A and B. β-Galactosidase assays of strains carrying the (A) PdegU–lacZ or (B) PaprE–lacZ transcriptional reporter fusion. Strains shown are WT (wild-type NRS4351), ΔmotB (NRS4345), motB + amyE::PIPTG-motB-D24A-lacI (NRS4397) grown in the absence or presence of 50 μM IPTG and ΔdegU (NRS4373).B. β-Galactosidase assays of strains carrying the PaprE–lacZ transcriptional reporter fusion. Strains shown are WT (wild-type NRS1561), ΔmotB (NRS3440), ΔmotB + amyE::PIPTG-motB-D24A-lacI (NRS3870) grown in the absence or presence of 50 μM IPTG. All cells in (A) and (B) were collected at the onset of stationary phase.C. Total protease activity assays performed with supernatants collected from cells grown in (B).Data in (A), (B) and (C) are plotted as the average of at least three independent replicates. In (C) data are represented as a fold change relative to the wild-type strain which was assigned value of 1. Error bars represent standard error of the mean. An asterisk denotes significance as calculated by the Student's t-test where * represents P < 0.05; **P < 0.01; and *** P < 0.001.D. Colony morphology of ΔmotB + amyE::PIPTG-motB-D24A-lacI (NRS3870) grown on LB agar in the absence and presence of 50 μM IPTG. SDS-PAGE analysis of γ-PGA collected from cultures of ΔmotB + amyE::PIPTG-motB-D24A-lacI (NRS3870) grown in the absence or presence of 50 μM IPTG.
