This work uses tiling microarrays to map S/MARs on Arabidopsis chromosome 4. S/MARs were found to be spaced more closely than in the large plant and animal genomes studied to date and preferentially enriched in poly(dA:dT) tracts, sequences that resist nucleosome formation. Most S/MARs occur near gene transcription start sites, and these genes show an increased probability of expression.
Abstract
Scaffold or matrix attachment regions (S/MARs) are found in all eukaryotes. The pattern of distribution and genomic context of S/MARs is thought to be important for processes such as chromatin organization and modulation of gene expression. Despite the importance of such processes, much is unknown about the large-scale distribution and sequence content of S/MARs in vivo. Here, we report the use of tiling microarrays to map 1358 S/MARs on Arabidopsis thaliana chromosome 4 (chr4). S/MARs occur throughout chr4, spaced much more closely than in the large plant and animal genomes that have been studied to date. Arabidopsis S/MARs can be divided into five clusters based on their association with other genomic features, suggesting a diversity of functions. While some Arabidopsis S/MARs may define structural domains, most occur near the transcription start sites of genes. Genes associated with these S/MARs have an increased probability of expression, which is particularly pronounced in the case of transcription factor genes. Analysis of sequence motifs and 6-mer enrichment patterns show that S/MARs are preferentially enriched in poly(dA:dT) tracts, sequences that resist nucleosome formation, and the majority of S/MARs contain at least one nucleosome-depleted region. This global view of S/MARs provides a framework to begin evaluating genome-scale models for S/MAR function.
INTRODUCTION
Early electron micrographic studies provided evidence that chromatin fibers form loops anchored to a proteinaceous nuclear matrix or scaffold (Berezney and Coffey, 1974; Paulson and Laemmli, 1977). These studies used preparations depleted of histones by extraction with either NaCl or lithium 3′-5′-diiodosalicylic acid (LIS). Histone removal disrupts nucleosomes and uncoils the DNA, forming loops that can be visualized by electron microscopy (Paulson and Laemmli, 1977). The loops are attached to the nuclear matrix at specific loci designated as scaffold/matrix attachment regions (S/MARs) (Cockerill and Garrard, 1986; Mirkovitch et al., 1988).
The nuclear matrix is a complex structure consisting of many different proteins (Capco et al., 1982; Calikowski et al., 2003) with multiple proposed functions (reviewed in Albrethsen et al., 2009; Simon and Wilson, 2011). Most of this work has been performed in animal systems, and less is known about the protein composition of the nuclear matrix in plants. Some plant nuclear matrix proteins have conserved roles, such as controlling chromosome dynamics (Lam et al., 2005), whereas others have roles in regulating development (Ng and Ito, 2010).
S/MARs are operationally defined as (1) endogenous DNA fragments that copurify with the nuclear matrix or (2) exogenous DNA fragments that show specific binding to the nuclear matrix in the presence of excess nonspecific competitor DNA (Cockerill and Garrard, 1986). S/MARs are variable in length, with bound fragment sizes ranging from 300 to 1000 bp (Gasser and Laemmli, 1986). Importantly, because the fragments are generally defined by restriction enzyme cuts, actual binding sites may constitute only a portion of the bound fragment.
Most S/MARs consist of AT-rich DNA (>70%) (Liebich et al., 2002), but the precise binding sites remain poorly defined in most cases, and sequence analysis has failed to reveal a strong consensus. One reason for this difficulty is that the nuclear matrix appears to recognize DNA structural features rather than primary sequence information (reviewed in Boulikas, 1993). Examples include a narrow minor groove associated with homopolymeric runs of dA:dT [poly(dA:dT) tracts] (Adachi et al., 1989; Käs et al., 1989; Rohs et al., 2009) and regions of strand unpairing (Bode et al., 1992), both of which are common features of AT-rich DNA. However, not all AT-rich DNA fragments bind to the nuclear matrix (von Kries et al., 1991; Dickinson et al., 1992; Morisawa et al., 2000). Sequence analysis is also complicated by the fact the fragments isolated as S/MARs often have flanking non-S/MAR sequence. Thus, it is not surprising that multiple attempts to develop predictive algorithms have been only partially successful in predicting S/MARs from sequence alone (reviewed in Evans et al., 2007).
S/MARs play critical roles in defining structural units of chromatin, functioning as boundary elements bordering regions of condensed or open chromatin structure (reviewed in Gerasimova et al., 2000), and some correlate with origins of DNA replication (Vaughn et al., 1990; Jenke et al., 2002; Mesner et al., 2003). In addition, S/MARs are often associated with promoters and genic regions, including introns where they may affect transcriptional activity or mRNA processing (Kumar et al., 2007; Alfonso-Parra and Maggert, 2010). Experiments with transgenes containing S/MARs have provided direct evidence of both positive and negative effects on transcription, with some S/MARs stabilizing transgene expression (reviewed in Allen et al., 2000).
Existing data are consistent with S/MARs playing a variety of roles, but a comprehensive understanding requires characterization of many more S/MARs across larger genomic regions. The largest region so far characterized in plants is 280 kb of the maize (Zea mays) genome (Avramova et al., 1995), where S/MARs appear to mark the boundaries of domains conserved between maize and sorghum (Sorghum bicolor). Recently, two groups presented large-scale analyses of S/MARs in the human genome, focusing on chromosomes 14 and 18 (Linnemann et al., 2009) or the 30-Mb portion in the initial ENCODE project (Keaton et al., 2011). Linnemann et al. (2009) distinguished two types of S/MARs depending on the method of histone extraction. S/MARs identified by NaCl extraction are often in silenced genes, while those identified by LIS extraction are frequently located near the 5′ region of genes producing transcripts. S/MARs identified by LIS extraction frequently map near transcription start sites (TSSs) and are preferentially associated with RNA polymerase II binding regions, expressed genes, and early replicating regions (Keaton et al., 2011).
We used a high-resolution tiling microarray to map 1358 S/MARs on chromosome 4 (chr4) of the model plant Arabidopsis thaliana, which has a compact 120-Mb genome. Arabidopsis S/MARs occur throughout chr4 with an average spacing of 11.4 kb, which is much closer than reported for the larger human and maize genomes (3.2 and 2.3 Gb, respectively). To assess potential functional diversity within the S/MAR population, we used k-means clustering to distinguish five groups of S/MARs based on their associations with other genomic elements. Each group was evaluated for nucleosome density, epigenetic modifications, and the occurrence of previously reported S/MAR sequence motifs. In addition, analysis of the S/MARs for all possible 6-mer sequence elements relative to the rest of chr4 uncovered an enrichment for poly(dA:dT) sequences, which are known to resist incorporation into nucleosomes. The majority of S/MARs overlap with at least one nucleosome-depleted region (NDR), but the pattern of poly(dA:dT) content of S/MARs is unique.
RESULTS
Identification of S/MARs
To identify Arabidopsis S/MARs, we adapted established procedures (Mirkovitch et al., 1984; Hall et al., 1991) to Arabidopsis suspension culture cells (Supplemental Figures 1 to 3). Isolated nuclei were extracted with buffer containing LIS to remove bound histones. The resulting “haloes” were digested with EcoRI and HindIII to release unbound DNA fragments, and DNA remaining with the matrix was isolated. This fraction included S/MARs as well as flanking sequences derived from the restriction fragments. S/MAR and total genome reference DNA were labeled with Cy3 or Cy5 and cohybridized to a NimbleGen custom tiling array covering chr4 with duplicate sets of 174,973 probes at a median tiling resolution of 100 bp. We did not attempt to map S/MARs in the heterochromatic knob and pericentromere because the resolution of our array is lower in these highly repetitive regions. Thus, when we refer to chr4 throughout this work, we are referring to the 15.4 Mb exclusive of the heterochromatic knob and pericentromeric DNA.
Peaks corresponding to S/MARs were identified with NimbleScan peak finder (Roche NimbleGen) using five different window sizes (see Methods). All putative S/MARs at a false discovery rate (FDR) ≤ 0.05 were retained, and peaks from all window sizes were merged. A total of 1358 S/MARs, ranging in size from 229 to 3666 bp, were identified covering 1,188,485 bp (7.7%) of the 15.4 Mb analyzed (Table 1; Supplemental Data Set 1). The mean length is 875 bp, and the broad distribution of sizes, including some S/MARs in the 200 bp range, are consistent with previous observations in animal systems (Bode et al., 1992; Frisch et al., 2002). However, all estimates include some flanking sequence along with the actual binding site(s) in any given S/MAR.
Table 1. Summary Information for S/MARs.
| Cluster | Count | Length (bp) | Coverage (%)a | AT Content (%) | ||
|---|---|---|---|---|---|---|
| Mean | Min | Max | ||||
| A | 424 | 864 | 230 | 2945 | 2.4 | 70.0 |
| B | 276 | 986 | 231 | 3666 | 1.8 | 72.9 |
| C | 285 | 941 | 232 | 2999 | 1.7 | 65.9 |
| D | 231 | 715 | 229 | 1963 | 1.1 | 58.7 |
| E | 142 | 821 | 328 | 1800 | 0.8 | 67.1 |
| Total | 1358 | 875 | 229 | 3666 | 7.7 | 67.5 |
Binding to the matrix as isolated in vivo is likely to be the most sensitive assay for matrix association. However, to further explore the specificity of binding, 31 putative S/MARs, along with eight non-S/MAR fragments that did not show matrix association in vivo, were tested in an in vitro binding assay (Hall et al., 1991) (Supplemental Figures 2 and 3 and Supplemental Data Set 2). PCR fragments derived from 26 of the 31 putative S/MARs (84%) exhibited specific binding in the presence of excess Escherichia coli DNA competitor. The five fragments that failed to bind under these conditions had lower AT contents relative to the other S/MARs and belonged to a specific class of S/MARs (see below and Supplemental Data Set 2). All of the PCR fragments amplified from regions not associated with the matrix as isolated also failed to bind to matrices in vitro.
Distribution of S/MARs on Chr4
Arabidopsis chr4 can be divided into regions based on cytology, the abundance of genes and transposable elements (TEs) (Figure 1), and replication time (Lee et al., 2010). We distinguished six regions along chr4: the distal short arm, the heterochromatic knob, the proximal short arm, the pericentromeric DNA, the proximal long arm, and the distal long arm. The distribution of the S/MARs is fairly uniform across chr4, ranging from a S/MAR every 11 kb in the distal long arm to every 12 kb in the proximal long arm (Figure 1; Supplemental Table 1). Previous reports for human cells showed that S/MARs are highly enriched in early replicating regions and depleted in late replicating regions (Keaton et al., 2011). In Arabidopsis, we observed only a modest 18% enrichment (P ≤ 0.001) of S/MARs in early replicating regions and a 19% depletion (P ≤ 0.001) in late replicating regions (Supplemental Figure 4).
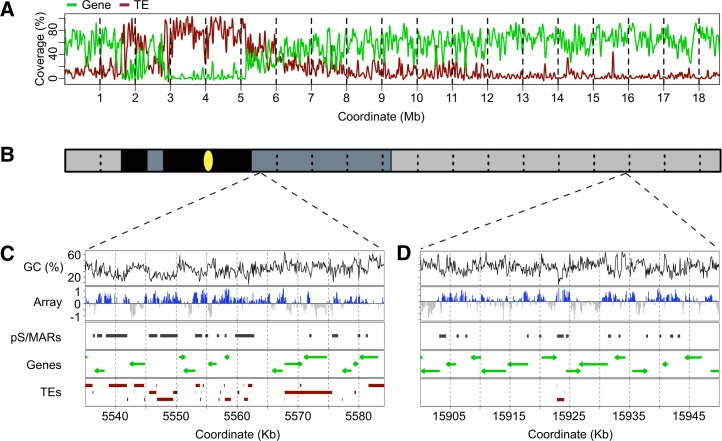
Figure 1.
Mapping of S/MARs in Select Regions of Arabidopsis Chr4.
(A) Gene and TE coverage for chr4. Chr4 was divided into 1-kb nonoverlapping bins, and the gene and TE coverage was calculated for each bin. This data was loess-smoothed in a 100-kb window for the plot.
(B) Chr4 can be subdivided into six regions based on the gene and TE coverage and the time of DNA replication (Lee et al., 2010). The position of the centromere is indicated by the yellow oval. The early-replicating distal long and short arms are the most euchromatic and are shaded light gray. The late-replicating proximal long and short arms have substantial heterochromatin and are shaded dark gray. The late-replicating constitutive heterochromatin of the knob and pericentromere are shaded black. S/MARs were not mapped in these last two regions.
(C) Mapping of S/MARs in a representative late-replicating region with significant TE content. Shown are the GC (%) and the log2 ratios for the S/MAR to genomic DNA microarrays with positive values highlighted in blue. Locations of computationally predicted S/MARs (pS/MARs) (Rudd et al., 2004) are shown for comparison. Genes and the direction of transcription are shown as green arrows, and TEs from TAIR10 Arabidopsis genome annotation are shown as brown boxes.
(D) Mapping of S/MARs in a representative early-replicating region. The mapped features are as described in (C).
Within chr4 regions, S/MARs are located in a variety of genomic contexts. In the TE-dense proximal long arm, S/MARs frequently associate with various TEs and TE fragments (Figure 1C). In the gene-dense distal long arm, S/MARs tend to occur in intergenic regions or closely flanking genes, although some S/MARs are entirely intragenic (Figure 1D). The distribution of our S/MARs is similar to the distribution of computationally predicted S/MARs (pS/MARs) for Arabidopsis chr4 (Frisch et al., 2002; Rudd et al., 2004) (Figures 1C and 1D). Of our 1358 S/MARs, 860 (63%) overlap with a pS/MAR (Supplemental Table 2). However, 498 S/MARs and 2032 pS/MARs do not overlap (Supplemental Table 2). For many of the latter, we observed weak or variable microarray signals that did not meet our FDR cutoff of ≤0.05 (Figures 1C and 1D). One interpretation of this result is that such regions may function as S/MARs only in a subset of cells. In regards to the 498 S/MARs that do not overlap with pS/MARs, we tested 10 examples in the in vitro binding assay, and all of them showed binding activity (Supplemental Data Set 2). This underscores the importance of in vivo mapping to fully characterize S/MARs.
Genomic Context of S/MARs
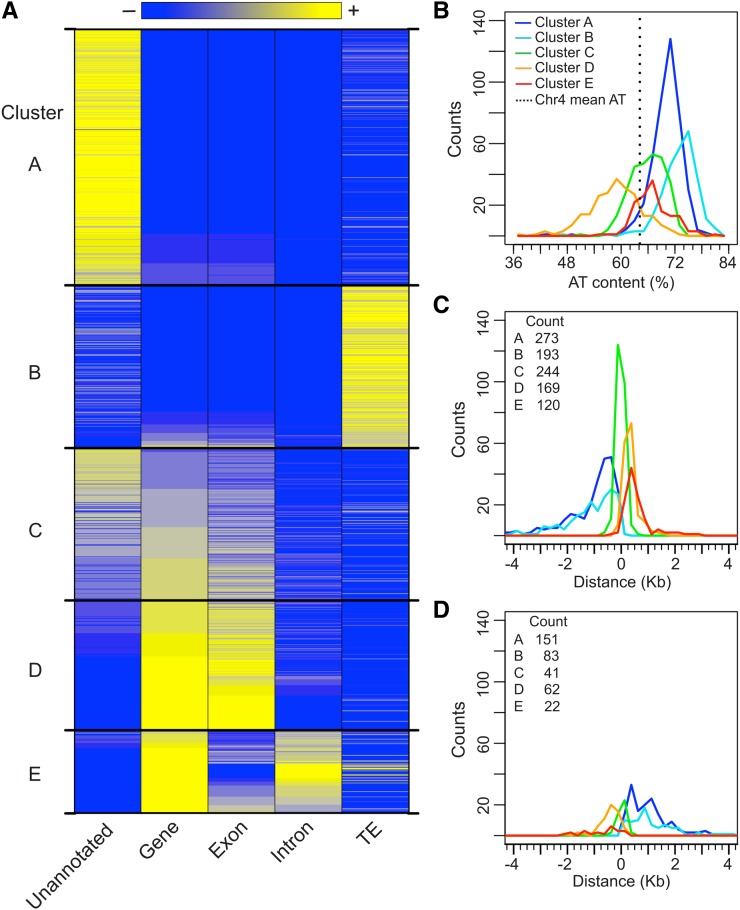
To determine the genomic context of S/MARs, we calculated the extent of intersection between the S/MARs and various genomic features in the Arabidopsis TAIR10 genome annotation (Buisine et al., 2008; Swarbreck et al., 2008). The intersection is the percentage of S/MAR sequence that overlaps with the following annotated features: genes, exons, introns, and TEs or TE fragments. We also determined the percentage of each S/MAR that has no annotation in TAIR10 as well as its AT content. K-means clustering of gene, exon, and TE content was used to group the S/MARs based on their genomic context. After testing a range of 2 to 25 clusters, we chose to use five clusters because it achieved good balance between biological relevance and the variance within the clusters (Figure 2A; Supplemental Figure 5 and Supplemental Table 3).
Figure 2.
Genomic Characteristics and Clustering of S/MARs.
(A) K-means clustering of S/MARs based on genomic context. The gene, exon, and TE coverage of each S/MAR was calculated and k-means clustering (k = 5) was performed. The results for each S/MAR within these five clusters are shown as a heat map, using 10% coverage increments. Unannotated and intron coverage is shown but was not used for clustering.
(B) Histograms of S/MAR AT content (%) for each cluster, using bins of 2%.
(C) Histograms of S/MAR position relative to the TSS of the closest gene. S/MARs were first grouped as either TSS- (or TTS)-proximal based on their absolute distance to the TSS or TTS of the nearest (or any overlapping) gene. Distances were calculated and corrected for gene strand so that S/MARs upstream of the TSS have negative values. Histograms were calculated using 250-bp bins, and the plot is restricted to the range ±4 kb (18 outliers are not shown). Also shown is the total count for each S/MAR cluster.
(D) Histograms of S/MAR position relative to the TTS of the closest gene. Distances were calculated as in (C). Two outliers are not shown. Also shown is the total count for each S/MAR cluster.
The properties of the resulting S/MAR clusters are summarized in Table 1 and Figure 2A. Cluster A S/MARs show minimal overlap with either genes or TEs and are located primarily in unannotated regions of chr4. Cluster B S/MARs have a strong association with TEs and minimal or no overlap with genes. Cluster C S/MARs have minimal TE content but approximately equal genic and unannotated content, suggesting that they flank genes. Cluster D S/MARs are associated with the exons of genes while Cluster E S/MARs are associated with the introns of genes. Clusters A and B have the highest AT content, while cluster D has the lowest (Table 1, Figure 2B). Interestingly, the majority of S/MARs that overlap with a pS/MAR are in clusters A and B. In cluster A, 310 of 424 S/MARs (73%) overlap with a pS/MARs, while 261 of 276 S/MARs (95%) in cluster B have a corresponding pS/MAR. Overlaps for Clusters C, D, and E are 60, 22, and 50%, respectively (Supplemental Table 2).
To better understand how S/MARs are positioned relative to genes, we determined the distance from each S/MAR midpoint to the closest annotated 5′ gene end or TSS and 3′ gene end or transcription termination site (TTS). Surprisingly, the majority of S/MARs are closer to a TSS than a TTS, with >80% of cluster C and E S/MARs proximal to a TSS. S/MARs in clusters A (66%) and B (70%), which show minimal overlap with genic sequences, are also closer to a TSS than a TTS (Figures 2C and 2D). For cluster C S/MARs, the association with a TSS is strikingly tight with a median distance of 22 bp upstream of the TSS.
Chromatin State of S/MARs and Flanking Regions
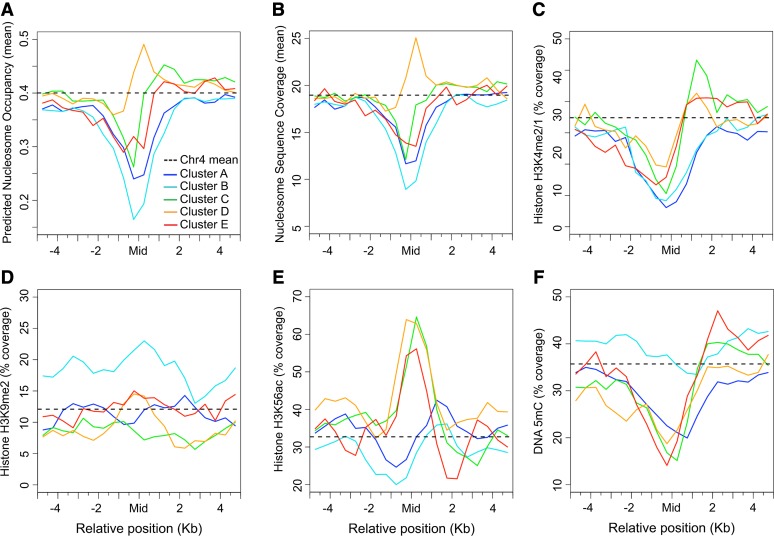
We next assessed the relationship between the S/MARs and their flanking sequences with selected chromatin features, including nucleosome density, histone modifications, and DNA methylation. We evaluated the nucleosome density using both predicted nucleosome occupancy values (Kaplan et al., 2009) and data from high-throughput sequencing of mononucleosomes isolated from Arabidopsis shoots (Chodavarapu et al., 2010). Additionally, we compared S/MAR positions to maps for three histone modifications, histone H3 lysine 4 mono- and dimethylation (H3K4me2/1), histone H3 lysine 9 dimethylation (H3K9me2), and histone H3 lysine 56 acetylation (H3K56ac) and to DNA methylation (DNA 5mC) maps. These maps were generated from the same cell line used for the S/MAR map (Tanurdzic et al., 2008).
The S/MARs were aligned at their midpoints and windows 5 kb upstream and downstream of the midpoints were delineated and subdivided into 20 500-bp bins. Any patterns common to the S/MARs should manifest at the midpoint bins, while patterns related to the genomic context of the S/MARs should appear in the flanking bins. Because of the close association between S/MARs and genes, upstream and downstream were defined based on the polarity on the gene most proximal to the midpoint of each S/MAR. The mean nucleosome occupancy or sequence coverage in the bins across the five S/MAR clusters was determined (Figures 3A and 3B). For the epigenetic modifications, the resolution of available data is inherently low, so the intersection of the S/MAR bins with DNA segments bearing each modification was calculated (Figures 3C to 3F).
Figure 3.
Chromatin Context of S/MARs by Cluster.
The colors shown in the key indicate the respective S/MAR clusters.
(A) Predicted average nucleosome occupancy scores based on the Arabidopsis DNA sequence (Kaplan et al., 2009) were used to estimate the probability of nucleosome occupancy of S/MARs and flanking regions. S/MARs were aligned at their midpoint and a window of ±5 kb was delineated and divided into 500-bp bins. Upstream and downstream were defined relative to the gene closest to each S/MAR. For each S/MAR, the mean predicted nucleosome occupancy in each bin was determined and then averaged for each S/MAR cluster.
(B) Similar to (A) but using experimental sequence coverage data for Arabidopsis shoot mononucleosomes (Chodavarapu et al., 2010). The units are the mean coverage for the sequence reads.
(C) to (F) Epigenetic modifications of S/MARs as shown described in (A) but using microarray results for chromatin immunoprecipitations to H3K4me2/1 (C), H3K9me2 (D), H3K56ac (E), and DNA 5mC (F) in our cell line (Tanurdzic et al., 2008). The microarray data were used to define segments of chr4 marked by each epigenetic modification. We then determined the coverage of these segments within each bin and expressed this as a percentage.
S/MAR clusters A, B, C, and E show a significant depletion of nucleosomes, both within the S/MARs and extending into the flanking regions (Figures 3A and 3B). For clusters A, C, and E S/MARs, this is not surprising given that these S/MARs are associated with intergenic, promoter and intronic regions, respectively, and these regions often have low nucleosome density (Cairns, 2009; Sun et al., 2009; Chodavarapu et al., 2010). For all S/MARs, there is a tendency for the downstream sequences to be nucleosome-enriched relative to the upstream sequences. This trend is a consequence of the close proximity between S/MARs and the TSS of genes and the fact that the mean length of chr4 Arabidopsis genes is quite short (e.g., only 2.3 kb or about one-fourth of our window size). Surprisingly, cluster B S/MARs have the lowest nucleosome occupancy (Figures 3A and 3B), even though they are characterized by TEs, which are often associated with repressive histone modifications such as H3K9me2 (Tanurdzic et al., 2008; Roudier et al., 2011). Cluster D S/MARs have both predicted nucleosome occupancy values and nucleosome sequence reads indicative of frequent incorporation into nucleosomes (Figures 3A and 3B). A possible explanation for this is that cluster D S/MARs may associate with the matrix in a subset of cells but not the entire population.
The epigenetic modifications associated with the S/MARs and flanking sequences are generally compatible with the nucleosome data. H3K4me2/1 is depleted in all S/MAR clusters (Figure 3C). This result is consistent with the fact that this modification is most prevalent in the body and TTS-proximal regions of Arabidopsis genes (Tanurdzic et al., 2008; reviewed in Liu et al., 2010) and our observation that S/MARs tend to be TSS-proximal (Figure 2C). The fact that we see a slight enrichment for H3K4me2/1 for S/MAR clusters C, D, and E downstream of the S/MAR midpoint (Figure 3C) is also consistent with this distribution.
Surprisingly, cluster B S/MARs are enriched for H3K9me2 (Figure 3D), even though nucleosome occupancy is reduced (Figures 3A and 3B). This apparent discrepancy may reflect the low resolution of the microarray data for H3K9me2 and the presence of flanking non-S/MAR sequences in the EcoRI-HindIII fragments used to map S/MARs. H3K9me2 is an abundant mark in Arabidopsis TEs, and it is likely that H3K9me2 in flanking TEs extends into adjacent sequences corresponding to S/MARs. However, we cannot rule out that, in a fraction of cells, cluster B S/MARs are packaged into H3K9me2-containing nucleosomes that do not bind the nuclear matrix.
For H3K56ac, we observed slight depletions in S/MAR clusters A and B but striking enrichment near the midpoints of S/MAR clusters C, D, and E (Figure 3E). For S/MAR clusters C and E, this is at odds with the nucleosome depletion (Figures 3A and 3B) and may reflect the same technical constraints mentioned above, given that H3K56ac is an abundant mark at the TSS of genes in Arabidopsis (Tardzic et al., 2008; Roudier et al., 2011). However, H3K56ac is enriched in regions where nucleosomes undergo turnover (Henikoff, 2008; Luger et al., 2012), and nucleosomes bearing this modification may be positionally unstable and not isolated in mononucleosomes. Consistent with this idea, H3K56ac is also enriched in cluster D S/MARs (Figure 3E). The positional instability of H3K56ac nucleosomes may allow the nuclear matrix to bind cluster D S/MARs and enhance the binding of cluster C and E S/MARs. Additionally, for clusters A and B, we observed a slight enrichment of H3K56ac downstream of the midpoint, while, for clusters C and E, there is a modest depletion (Figure 3E). This is expected because, on average, sequences downstream of a cluster A or B S/MARs are increasingly likely to be genic, whereas for cluster C, D, and E S/MARs the downstream sequences are increasingly likely to be intergenic (Figure 3E).
Finally, we observed modest depletions of DNA 5mC in S/MAR clusters A, C, D, and E, while cluster B S/MARs were neither enriched nor depleted (Figure 3F). All S/MAR clusters except D are AT-rich (Figure 2B), so it was not surprising that DNA 5mC is reduced. While cluster D S/MARs are relatively GC-rich (Figure 2B), they are TSS-proximal (Figure 2C), and DNA 5mC tends to be reduced near the TSS of Arabidopsis genes (Tanurdzic et al., 2008; Zhang et al., 2009a; Roudier et al., 2011). For cluster B S/MARs, these AT-rich sequences (Figures 2A and 2B) are proximal to TEs, and TEs are usually hypermethylated (Tanurdzic et al., 2008; Roudier et al., 2011), so the reduced GC content and hypermethylation of flanking sequencing may be compensating for each other in the analysis. Again, due to the close association of S/MARs to the TSS of genes, there is a tendency for downstream sequences to show increased 5mC relative to the upstream sequences (Figure 3F).
In summary, S/MAR clusters display nucleosome occupancy values and associated epigenetic modifications consistent with their AT content and genomic context. The sequences flanking S/MARs show epigenetic patterns consistent with the tight association between S/MARs and the TSS of genes. However, we did not observe patterns in the S/MAR flanking regions that reflect the exon-intron structure of genes. A likely explanation for this is that Arabidopsis introns tend to be quite small (160 bp for chr4) relative to our 500-bp bin size, and introns and exons associated with different S/MARs are unlikely to align in these bins. The most interesting result is the striking enrichment of H3K56ac in S/MAR clusters C, D, and E. This histone modification is associated with nucleosomes undergoing turnover, which may explain why some of these sequences are accessible and bind to the nuclear matrix.
S/MARs and TEs
The 276 S/MARs in cluster B are associated almost exclusively with TEs (Figure 2A), and an additional 271 S/MARs have at least some association with TEs (Supplemental Table 1). Previous studies showed that certain S/MARs in Drosophila melanogaster (Nabirochkin et al., 1998), human cells (Rollini et al., 1999), and plants (Avramova et al., 1998; Tikhonov et al., 2000) contain sequences derived from various TEs. In addition, a survey for the presence of TEs in a collection of human S/MARs found an enrichment of TE-derived sequences (Jordan et al., 2003). In these cases, the statistical significance of the apparent enrichment of TE sequences in S/MARs could not be evaluated because of the small sample sizes.
To estimate the statistical significance of the association between S/MARs and each of the 18 annotated TE superfamilies (TE-SFs) (Buisine et al., 2008; Swarbreck et al., 2008), we determined the percent composition of each TE-SF in the S/MARs and in chr4 (Supplemental Figure 6A and Supplemental Table 4). The enrichment of TE-SFs is expressed as the ratio of TE-SF composition in S/MARs to the composition in chr4. We found a wide range of ratios from 0.13 for LTR/Copia elements to 5.0 for DNA/Mariner elements (Supplemental Table 4). To estimate P values, we generated a null distribution from 100,000 permuted S/MAR sample sets in which the start coordinates were randomly selected along chr4. This analysis revealed that the S/MARs are significantly enriched for sequences derived from five TE-SFs: DNA, DNA/Harbinger, DNA/Mariner, DNA/MuDR, and RC/Helitron (Supplemental Table 4). Only sequences in the LTR/Copia TE-SF are depleted in S/MARs (Supplemental Table 4).
Given the high AT content of cluster B S/MARs (Figures 2A and 2B), the observed enrichment of certain TE-SFs in S/MARs may reflect their AT content. To test this hypothesis, we calculated the AT content of each of the TE-SFs and used a simple linear model to evaluate the relationship between TE-SF AT content and S/MAR enrichment. We observed a strong positive correlation (R2 = 0.56, P value = 0.0003) for TE-SF AT content and S/MAR enrichment (Supplemental Figure 6B). All of the AT-rich TE-SFs show at least some degree of enrichment in S/MARs, although the enrichment was modest or not statistically significant for DNA/Tc and DNA/Pogo (Supplemental Figure 6B and Supplemental Table 4). Finally, while this relationship between certain TE-SFs and S/MARs is interesting, it is important to note that the majority of our S/MARs (811) have no significant TE content (Figure 2A).
S/MARs and Gene Expression
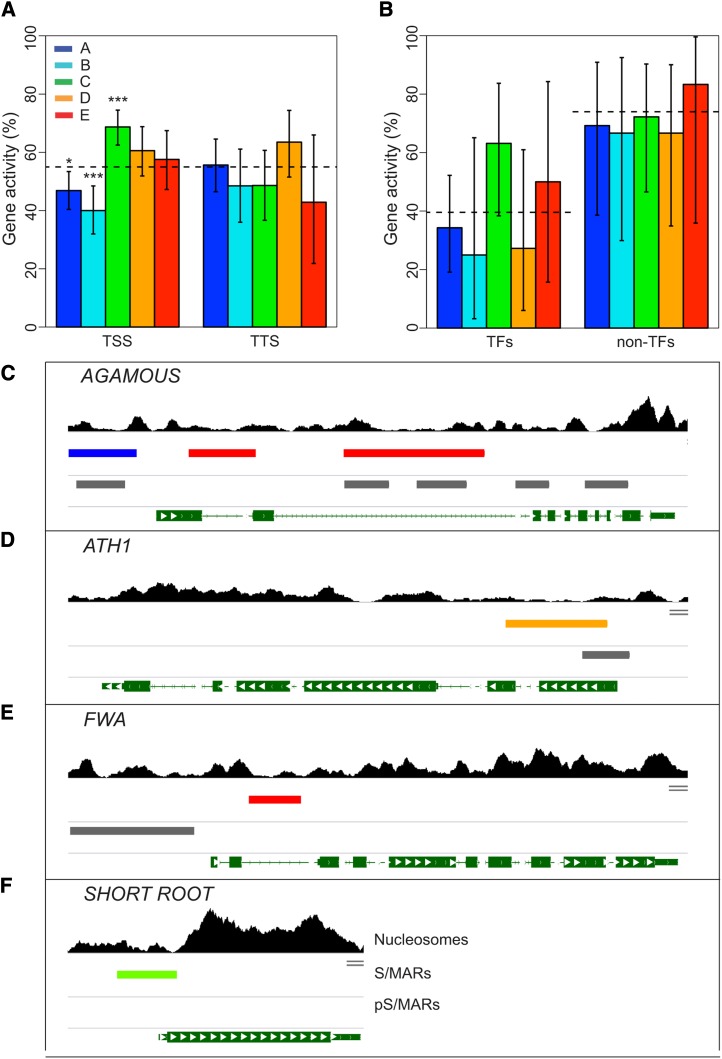
Over 60% of our mapped S/MARs (828 in total) overlap with annotated genes (excluding TE genes) (Figure 2A; Supplemental Data Set 1). Additionally, the S/MARs tend to associate preferentially with the TSS of annotated genes and not the TTS (Figures 2C and 2D). S/MARs in transgene constructs can increase expression in plants (Allen et al., 2000; Butaye et al., 2004; Thompson et al., 2007) and have been implicated in gene regulation in both plants and animals (Tetko et al., 2006; Ottaviani et al., 2008; Linnemann et al., 2009; Ng et al., 2009; Keaton et al., 2011). These studies and the tight association between our S/MARs and the TSS of genes prompted us to test for a correlation between S/MAR proximity and gene activity.
To accomplish this, all genes that either overlap (911) or immediately precede or follow a S/MAR (795) were identified, resulting in a list of 1706 genes (Supplemental Data Set 3). The remaining 2640 genes on chr4 are flanked only by other genes. The S/MAR-associated genes were categorized based on whether the nearest S/MAR is proximal to the TSS or TTS and by S/MAR cluster (Supplemental Data Set 3). As a metric for gene activity, MAS5 presence/absence calls (Hubbell et al., 2002) from Affymetrix expression data for our cell line were used (Tanurdzic et al., 2008). Gene expression values were avoided because these likely reflect mRNA stability as much as transcriptional activity. The number of expressed genes in each category was tabulated, excluding genes not on the microarray, and the binomial distribution was used to assess the statistical significance with the overall activity of chr4 genes as the reference.
We found that genes with cluster A or B S/MARs upstream of the TSS are less likely to be expressed than the control (P = 1.3 × 10−2 and P = 3.1 × 10−4, respectively), whereas genes with cluster C S/MARs near the TSS are more likely to be expressed (P = 1.4 × 10−5) (Figure 4A; Supplemental Table 5). By contrast, there was no significant correlation between gene activity and the presence of a TTS-proximal S/MAR (Figure 4A; Supplemental Table 5). Together, these results suggested that the proximity of the S/MAR to the gene promoter influences gene expression, and cluster C S/MARs have the largest effect, possibly because they have the tightest association with promoters (Figure 2C). The apparent repression of gene expression for TSS-proximal cluster A and B S/MARs may not be direct, although there are examples of S/MARs distal to the TSS having repressive activity (Tetko et al., 2006; Ng et al., 2009; Dinh et al., 2012). Interestingly, the presence of intragenic S/MARs in clusters D and E did not significantly impact gene activity in this study, although previous studies indicated that intragenic S/MARs are repressive (Tetko et al., 2006; Linnemann et al., 2009; Ng et al., 2009; Dinh et al., 2012). Our results suggest that different S/MARs can facilitate or repress gene expression.
Figure 4.
Correlation of S/MARs with Gene Activity.
The colors shown in the key indicate the respective S/MAR clusters.
(A) All genes that either overlap or are adjacent to an S/MAR were identified and categorized as having either TSS- or TTS-proximal S/MARs based on the location of the closest S/MAR midpoint. Gene activity was determined from MAS5 presence/absence calls from Affymetrix expression experiments for our cell line (Tanurdzic et al., 2008) and is the percentage of genes with detectable mRNA in each category. Totals for the TSS-proximal S/MARs are 239, 145, 243, 137, and 99 for S/MAR clusters A, B, C, D, and E, respectively, and 124, 66, 72, 74, and 21 for S/MAR clusters A, B, C, D, and E, respectively, for the TTS-proximal S/MARs. The dashed line shows the mean for all chr4 genes, and the error bars indicate the 95% confidence interval from the binomial test, and P value significance cutoffs are indicated as follows: *P ≤ 0.05, **P ≤ 0.001, and ***P ≤ 0.0001.
(B) Gene activity for TF genes and non-TF genes in the functional cluster. We identified all transcription associated genes on chr4 based on the enriched GO terms in the identified functional cluster and grouped them as TFs and non-TFs based on the AtTFDB (Palaniswamy et al., 2006). Activity of the transcription-associated genes with no TSS-proximal S/MAR is shown as a dashed line. The total TF genes are 35, 8, 19, 11, and 8 for S/MAR clusters A, B, C, D, and E, respectively, and 13, 9, 18, 12, and 6 for S/MAR clusters A, B, C, D, and E, respectively, for the non-TF genes. The error bars indicate the 95% confidence interval from the binomial test.
(C) to (F) Developmentally important TF genes are associated with S/MARs. Shown are AGAMOUS (C), ATH1 (D), FWA (E), and SHORT ROOT (F) with the S/MARs and pS/MARs indicated. The tracks (top to bottom) are mononucleosome sequence coverage as in Figure 3B. The locations of the S/MARs color-coded by cluster (described above). The locations of the computationally predicted S/MARs (pS/MARs) (Rudd et al., 2004) are shown in gray for comparison. Genes are shown as green segments with exons indicated by boxes and the direction of transcription by white arrows.
S/MARs and Genes Associated with Transcription
To examine the relationship between the genes with a TSS-proximal S/MAR, we used the functional annotation tool at the DAVID Bioinformatics Database (Huang et al., 2009a) using chr4 genes as the background. Strikingly, only one functional group of 167 genes was strongly associated with S/MARs (Table 2; Supplemental Data Set 4). This group was enriched for Gene Ontology (GO) terms and Protein Information Resource keywords associated with transcription (Table 2; Supplemental Data Set 4). To confirm this result, a list of transcription factors (TFs) from the Arabidopsis Gene Regulatory Information Server Transcription Factor Database (AGRIS AtTFDB) was used (Palaniswamy et al., 2006). This analysis showed that 103 of the 250 TFs on chr4 have a TSS-proximal S/MAR. Ninety-two of these TF genes were already identified with DAVID (Table 2). We did not include the additional 11 putative TFs in our final list because the annotation in the AtTFDB is inconclusive for these genes. The remaining 75 non-TF genes in the functional group identified by DAVID are diverse (Supplemental Data Set 4) and include genes of unknown function and genes encoding protein kinases that act in the nucleus, transcriptional repressors, and proteins with AT hook motifs. Interestingly, AHL1, which has an AT hook motif, encodes a S/MAR binding protein (Fujimoto et al., 2004).
Table 2. Annotation for Functional Cluster of Genes with a TSS-Associated S/MAR.
| Annotationa | Aspect | Term/Keyword | Countb | P Valuec |
|---|---|---|---|---|
| GO Terms | Biological Process | Regulation of transcription | 116 | 1.7 × 10−2 |
| Transcription | 86 | 1.2 × 10−2 | ||
| Regulation of transcription, DNA-dependent | 77 | 2.8 × 10−3 | ||
| Regulation of RNA metabolic process | 77 | 2.8 × 10−3 | ||
| Molecular Function | DNA binding | 134 | 5.2 × 10−3 | |
| Transcription regulator activity | 121 | 5.6 × 10−3 | ||
| TF activity | 107 | 2.8 × 10−3 | ||
| Sequence-specific DNA binding | 37 | 1.2 × 10−1 | ||
| PIR Keywords | Nucleus | 108 | 2.9 × 10−3 | |
| DNA binding | 82 | 2.8 × 10−3 | ||
| Transcription regulation | 81 | 3.5 × 10−4 | ||
| Transcription | 80 | 4.9 × 10−4 | ||
| AtTFDB | Arabidopsis TFs | 92 | 2.3 × 10−4 |
The functional annotation of the 1165 genes with a TSS-associated S/MAR were analyzed for overrepresented GO terms and Protein Information Resource (PIR) keywords using the DAVID functional annotation tool with default settings (Huang et al., 2009a). One significantly enriched cluster of 167 genes was identified.
GO terms and PIR keywords are from the DAVID database (Huang da et al., 2009a). AtTFDB is the Arabidopsis Transcription Factor Database (Palaniswamy et al., 2006).
Eleven putative TFs were removed from the count for AtTFDB because of insufficient supporting annotation.
We next compared the activities of genes in the transcription functional group that are associated with a TSS-proximal S/MAR with the activities of similar genes that are not associated with a TSS-proximal S/MAR (Supplemental Data Set 5). Separate comparisons were made for TF and non-TF genes, based on their GO and AtTFDB designations. This comparison revealed that TF genes are less likely to be expressed than non-TF genes in this functional group (Figure 4B). More importantly, the impact of the different S/MAR clusters on TF gene expression was similar to their impact on expression in the original list of 1165 genes with a TSS-proximal S/MAR (compared with Figure 4A). Strikingly, 63% (12 of 19) of the TF genes with a TSS-proximal cluster C S/MAR are active compared with 40% (36 of 91) of those without a TSS-proximal S/MAR (Figure 4B). Even though this difference in gene activity was much greater for TF genes as opposed to chr4 genes in general (Figure 4B), it was only modestly significant (P = 0.06) (Supplemental Table 5). The presence of a cluster C S/MAR had no apparent effect on the activity of the non-TF genes (Figure 4B).
We mapped S/MARs to several TF genes that play key roles in plant development, including AGAMOUS, ATH1, FWA, and SHORT-ROOT (Figures 4C to 4F). Interestingly, the AGAMOUS gene also has a S/MAR in the second intron, which is critical for proper gene regulation (Sieburth and Meyerowitz, 1997; Deyholos and Sieburth, 2000; Hong et al., 2003; Dinh et al., 2012). Earlier studies also predicted and/or mapped S/MARs in the AGAMOUS and ATH1 genes (van Drunen et al., 1997; Rudd et al., 2004). The association of S/MARs with TF genes may provide an additional layer of regulation mediated by binding to the nuclear matrix.
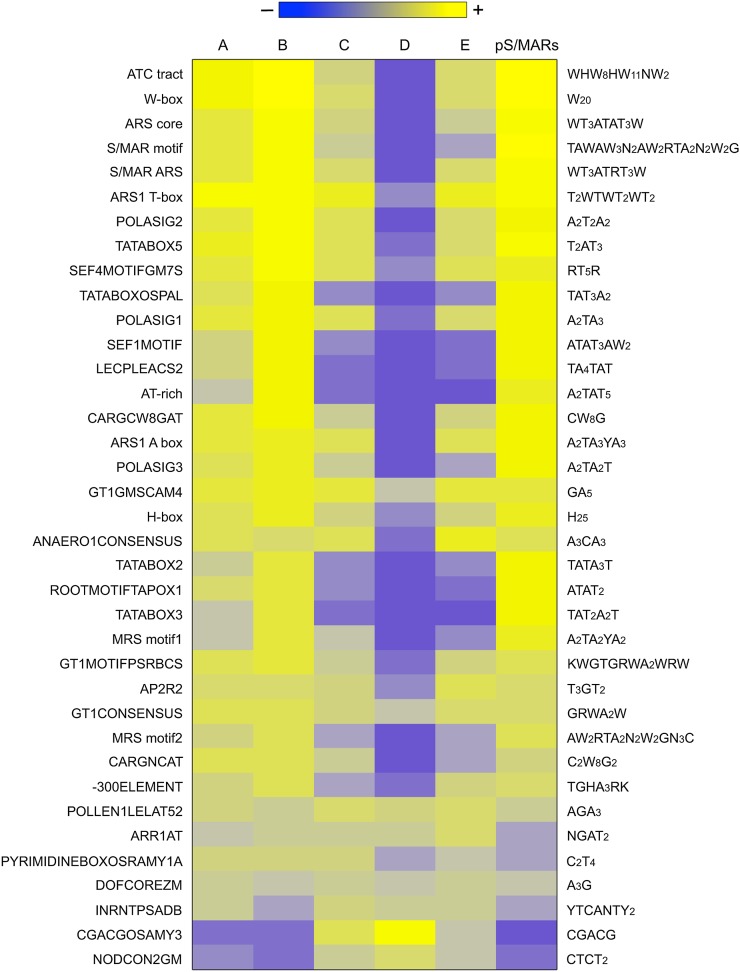
DNA Motifs Associated with S/MARS
Certain DNA sequence motifs are thought to be associated with S/MARs. Most of these motifs fall into two extremes, short and specific or long and degenerate, both of which tend to be AT-rich (Supplemental Data Set 6). In general, specific cis-elements have not been associated with S/MARs, although an enrichment of S/MARs in regulatory factor binding regions was shown in human cells (Keaton et al., 2011). The tight association of S/MARs with the TSS of genes (Figure 2C) suggested that specific cis-elements might contribute to S/MAR function. Therefore, we analyzed the S/MARs for possible overrepresentation of 456 annotated plant cis-elements (Higo et al., 1999) as well as 22 S/MAR-specific motifs. A putative binding site for the TF APETALA2 (AP2) that occurs in the S/MAR located in the second intron of AGAMOUS (Dinh et al., 2012) was also included, for a total of 479 specific DNA motifs (Supplemental Data Set 6).
To assess enrichment, the density of each motif (expressed as counts per kilobase) on chr4 and in each S/MAR (Supplemental Data Sets 6 and 7) was calculated. The pS/MARs were also included in this analysis for comparison. t tests were then used to determine if the density of the motifs is higher in the S/MAR clusters compared with chr4, correcting for multiple comparisons. Forty-five motifs are significantly enriched in at least one S/MAR cluster, with enrichment values ranging from a minimum of 11% to a maximum of 300% (Supplemental Data Set 8). Of these, 37 are enriched in clusters A, B, C, or E S/MARs and are shown in Figure 5. Only 11 of the 22 putative S/MAR motifs are enriched, but 26 putative cis-elements also show enrichment (Figure 5; Supplemental Data Set 8). Many of these cis-elements are similar to the S/MAR motifs and include short AT-rich motifs, such as TATA boxes, polyadenylation signals, and the AP2 binding site (Figure 5; Supplemental Data Set 8). There are also several long and degenerate AT-rich motifs, including two CArG boxes that are binding sites for MADS family TFs, such as AGAMOUS (Sieburth and Meyerowitz, 1997; Hong et al., 2003). Not surprisingly, the enriched motifs are similar in S/MAR clusters A, B, C, and E, whereas those in cluster D are distinct (Supplemental Data Set 8). The motifs specific to cluster D S/MARs have a higher GC content, but there is no clear sequence relationship among them.
Figure 5.
Analysis of DNA-Motif Content of S/MARs.
List of 479 sequence motifs was assembled including S/MAR-specific motifs and annotated cis-elements (Supplemental Table 11). Forty-five motifs were significantly overrepresented in at least one S/MAR cluster as determined by t tests of motif density in S/MARs relative to the density of the motif in chr4 (Supplemental Table 12). Shown is the percentage of enrichment for 37 motifs that were enriched in S/MAR clusters A, B, C, and E. Eight motifs that were enriched only in cluster D S/MARs are not shown. Motif references are included in Supplemental Table 13.
We observed several general trends for the motifs associated with cluster A, B, C, and E S/MARs. First, most are AT-rich (Figure 5). Second, degenerate motifs are more enriched than specific motifs, especially in AT-rich motifs that allow either an A or T at multiple positions (e.g., W-boxes). Third, most of the S/MARs contain multiple motifs and/or multiple copies of a single motif (Supplemental Data Set 7). This redundancy suggests that S/MAR binding is attributable to a general feature of DNA sequence rather than a specific sequence motif, analogous to nucleosome binding preferences (Kaplan et al., 2009; Zhang et al., 2009b).
Abundance of 6-Mers in S/MARs
Liebich et al. (2002) suggested that certain AT-rich 6-mers are overrepresented in S/MARs, but the small number of S/MAR sequences then available limited their conclusions. Our data set, consisting of 1358 in vivo–mapped S/MARs from a single organism, allows a more comprehensive analysis.
We determined the frequency of the 2080 nonredundant 6-mers in the S/MARs and across chr4. To determine the statistical significance of any observed enrichment, the binomial distribution was used to calculate P values, using the 6-mer abundance on chr4 as a reference. In addition to the S/MARs, Arabidopsis pS/MARs (Rudd et al., 2004), NDRs from Arabidopsis shoots (Chodavarapu et al., 2010), as well as promoters, exons, and introns for chr4 genes and annotated chr4 TEs were also included.
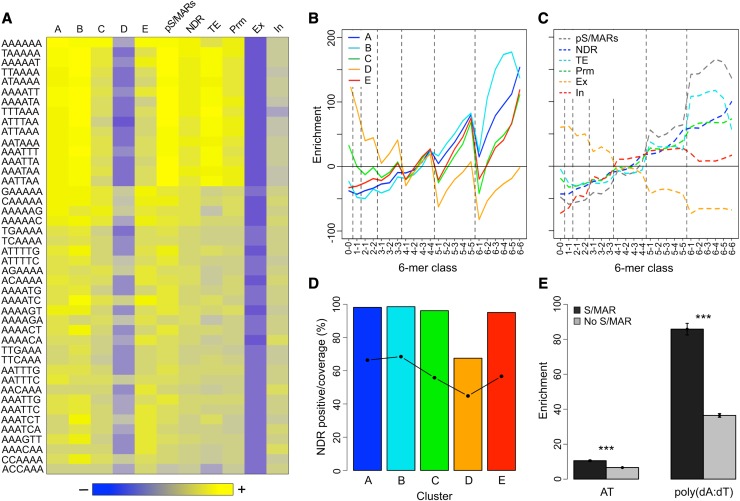
This analysis uncovered 510 6-mers that are enriched in S/MAR clusters A, B, C, D, and E collectively (Supplemental Data Set 9). However, a comparison of the enriched 6-mers across the five S/MAR clusters showed that no 6-mers are enriched in all the clusters (Supplemental Data Set 9). This was due entirely to the inclusion of cluster D. Of the 297 6-mers enriched in cluster D, 233 are enriched only in cluster D (Supplemental Data Set 9). When cluster D was excluded from the analysis, 44 6-mers are enriched in the four remaining S/MAR clusters, the NDRs, and promoters, and 43, 42, and 40 are also enriched in the pS/MARs, TEs, and introns, respectively (Figure 6A). None of the 44 6-mers are enriched in exons (Figure 6A). Twenty-three of these 6-mers are also among the 34 most abundant 6-mers in the S/MARs examined by Liebich et al. (2002).
Figure 6.
Analysis of the 6-Mer Content of S/MARs and Associated Regions.
(A) The 6-mer content of S/MARs and select genomic regions. The occurrence of the 2080 nonredundant 6-mers was determined for each S/MAR cluster, pS/MARs, NDRs, promoters (Pr), exons (Ex), introns (In), and TEs on chr4. The heat map shows enrichment data for the 44 overrepresented 6-mers that are in common between S/MAR clusters A, B, C, D, and E. The heat map is scaled so that blue indicates depletion, gray is no enrichment or depletion, and yellow indicates enrichment. The maximum depletion shown is 73%, while the maximum enrichment is 230%.
(B) Enrichment of 6-mer classes as a function of their poly(dA:dT) content for the five S/MAR clusters. All 6-mers were classified based on their poly(dA:dT) content, as described in the text. The dashed lines are included to group the 6-mers by class.
(C) As in (B) but for the following genomic regions: pS/MARs, NDRs, TEs, promoters, exons, and introns.
(D) Percentage of S/MARs by cluster that overlap with a NDR and the NDR sequence coverage for these S/MARs. The percentage of NDR-positive S/MARs is shown as bars, while the NDR sequence coverage is shown by the points and solid line.
(E) Comparison of the enrichment for AT and poly(dA:dT) content of S/MAR-positive and -negative NDRs. NDRs were partitioned into S/MAR-positive and -negative groups, and the AT content relative to chr4 was determined. For poly(dA:dT) content, sequence motifs comprising four or more contiguous As or contiguous Ts were identified for chr4 and for the NDRs, and the poly(dA:dT) enrichment for the NDRs, relative to chr4 was calculated. A Welch two-sample t test was used to compare the NDR groups. The error bars show the 95% confidence intervals for the means. For both comparisons, the difference between the means was highly significant (P ≤ 10−16) as indicated by the asterisks (Supplemental Table 16).
The most enriched 6-mers in S/MARs, with the exception of cluster D, are AT rich (Figure 6A). Cluster D S/MARs are depleted for many of these AT-rich 6-mers and, instead, have elevated levels of GC-rich 6-mers (Figure 6A; Supplemental Data Set 9). However, when compared with exons, cluster D S/MARs display a modest enrichment for some AT-rich 6-mers (Figure 6A), suggesting that the actual matrix binding site may be a minor constituent of these regions. Clusters C and E S/MARs display a pattern that is intermediate between clusters A and B and cluster D, consistent with their genomic context (Figure 2A). Cluster E S/MARs, which have considerable intron content (Figure 2A), have a 6-mer enrichment pattern that is distinct from chr4 introns (Figure 6A). There is a strong correlation between the enrichment of a particular 6-mer in NDRs and in all S/MARs except those in cluster D (Figure 6A). The 6-mer patterns of cluster B S/MARs, pS/MARs, and TEs is very similar, suggesting that the computer algorithm used to predict the pS/MARs favored a subset of sequences (Frisch et al., 2002; Rudd et al., 2004).
In addition to the AT content of the 6-mer, the number of contiguous A or contiguous T residues, or poly(dA:dT) content, appears to be important. Forty-three of the 44 common 6-mers have a minimum poly(dA:dT) content of three, including all seven 6-mers with a poly(dA:dT) content of five or six (Figure 6A). Poly(dA:dT) tracts have previously been implicated in S/MAR function (Adachi et al., 1989; Käs et al., 1989; Liebich et al., 2002). To examine this in more detail, we classified the 6-mers based on their AT and poly(dA:dT) content (i.e., 6-6, 6-5, 6-4, …, 2-2, 2-1, 1-1, 0-0) and determined the mean enrichment of these 6-mer classes in each type of sequence (Figure 6B). There is a striking pattern of 6-mer enrichment in S/MARs that is dependent on both AT and poly(dA:dT) content. With the sole exception of AAAAAA/TTTTTT in cluster B S/MARs, for any given AT content the more enriched 6-mers have higher poly(dA:dT) content. This pattern of enrichment leads to a characteristic “saw tooth” pattern in Figure 6B. This pattern is especially prominent for S/MAR clusters A, B, C, and E, but it can also be seen for cluster D S/MARs, where AT-rich 6-mers are actually depleted relative to chr4. Comparing the pattern of enrichment for cluster D S/MARs to the pattern for exons reveals that they are remarkably distinct. In fact, none of the other regions that we examined display this pattern, including the pS/MARs (Figure 6C).
Thus, a signature sequence for S/MAR binding is high poly(dA:dT) content. Intriguingly, poly(dA:dT) stretches are thought to resist incorporation into nucleosomes because of their unique biophysical properties (Segal and Widom, 2009). In addition, poly(dA:dT) stretches have been implicated in the modulation of both gene expression and DNA replication, probably through their ability to disrupt nucleosome arrays (Anderson and Widom, 2001; Field et al., 2008; Segal and Widom, 2009; Raveh-Sadka et al., 2012). Hence, it is not surprising that the NDRs are also rich in poly(dA:dT) tracts (Figure 6B).
The vast majority of S/MARs (92%) overlap with at least one NDR (Figure 6D; Supplemental Table 6), and even the majority (68%) of cluster D S/MARs contain a NDR (Figure 6D; Supplemental Table 6). For S/MARs that overlap with a NDR, the NDRs account for much of the S/MAR sequence ranging from 45% for cluster D S/MARs to 68% for cluster B S/MARs (Figure 6D; Supplemental Table 6). Importantly, all of the S/MARs that bound in vitro, with the exception of two cluster D S/MARs, overlapped with at least one NDR, and the five cluster D S/MARs that did not bind, lacked a NDR.
To determine if the NDRs located in S/MARs were different from typical NDRs, the NDRs were partitioned into S/MAR-positive (2407) and S/MAR-negative groups (12,677), and the enrichments of AT content and poly(dA:dT) content relative to chr4 were compared using a t test. For poly(dA:dT) content, we identified all sequence motifs with four or more contiguous As or contiguous Ts. Strikingly, the S/MAR-positive NDRs are more enriched for AT content (P ≤ 1 × 10−16), and especially for poly(dA:dT) content (P ≤ 1 × 10−16), than S/MAR-negative NDRs (Figure 6E; Supplemental Table 7). These atypical NDRs located in S/MARs likely contain the actual matrix binding sites of the larger fragments isolated as S/MARs.
DISCUSSION
We mapped 1358 S/MARs in the 15.4-Mb euchromatic regions of Arabidopsis chr4. S/MARs are distributed along the entire chromosome, and their distribution is largely independent of gene and TE density. The average spacing is 11.4 kb, similar to previous small-scale mapping studies that reported spacings as short as 5 kb (van Drunen et al., 1997; Tachiki et al., 2009). The relatively short distances between S/MARs in Arabidopsis contrast with chromosome-scale analyses in the much larger human genome, where average spacing ranges from 44 to 88 kb (Linnemann et al., 2009; Keaton et al., 2011). This disparity may reflect differences in genome size, repeat content, and gene density, given that the Arabidopsis genome is <5% as large as the human genome but has a similar number of genes. This idea is supported by a comparison of S/MARs associated with four genes near the ADH1 locus in maize and sorghum, which showed that the relative positions of S/MARs are conserved but TE insertions increase their absolute spacing in the larger maize genome (Tikhonov et al., 2000).
An association between TE-derived sequences and S/MARs has been shown for many model organisms (Avramova et al., 1998; Tikhonov et al., 2000; Byrd and Corces, 2003; Jordan et al., 2003). We found that chr4 S/MARs overlap with sequences derived from annotated Arabidopsis TE-SFs (Supplemental Figure 6 and Supplemental Table 4). This association was not statistically significant for many TE-SFs. However, there was specific enrichment for sequences derived from the Mariner, Harbinger, and MuDR superfamilies of DNA transposons as well as for Helitrons. The sequences of these TE-SFs are all AT rich, providing a likely explanation for their enrichment in S/MARs. The existence of TE-S/MAR associations raises questions about the relationship between matrix binding and transposition potential. This type of analysis is beyond the scope of our experiments, but our results lend support to reports postulating that TEs can contribute to genome organization patterns by moving S/MARs to new locations (Tikhonov et al., 2000; reviewed in Bennetzen, 2000).
We used k-means clustering to divide Arabidopsis S/MARs into five clusters based on their locations in relation to other genomic features. The clusters differ with respect to epigenetic marks, nucleosome occupancy, sequence characteristics, and the likelihood of expression of closely associated genes. While this clustering proved useful, our 6-mer motif analysis suggested that the matrix binding sites buried within the S/MARs are similar between clusters and that the clustering is largely driven by the flanking sequence. Nevertheless, the clustering of S/MARs suggests that they might have different biological functions depending on their context.
S/MARs are thought to have both structural and regulatory roles. For example, S/MARs in intergenic regions (clusters A and B) may delineate chromatin domains, while S/MARs near or in genes (clusters C, D, and E) may potentiate or inhibit expression (Avramova et al., 1998; Tikhonov et al., 2000; Byrd and Corces, 2003; Jordan et al., 2003). However, because distances from the TSS tend to be small, and their distribution continuous, distinctions between structural and regulatory S/MARs may be more arbitrary in Arabidopsis than in organisms with larger genomes.
Regulatory S/MARs have been associated with positive and negative effects on gene expression. S/MARs can promote or stabilize expression of nearby transgene(s) (Allen et al., 1996; Butaye et al., 2004; reviewed in Allen et al., 2000; Thompson et al., 2007). In animal cells, native S/MARs are often located near or within enhancers or promoters of genes (Keaton et al., 2011), and some genes require a S/MAR near their 5′ends for proper expression (Gasser and Laemmli, 1986; Ng et al., 2009). Arabidopsis S/MARs are frequently located at the TSS of genes (Figure 2C), and genes associated with a cluster C S/MAR are more likely to be expressed (Figure 4A). Interestingly, the effect of S/MARs on the likelihood of expression is more striking for genes encoding TFs (Figure 4B). An earlier report also showed a significant association of S/MARs with TF genes in Arabidopsis, although that study relied on S/MARs identified solely in silico (Tetko et al., 2006).
The ability of S/MARs to facilitate gene regulation is likely linked to their strong association with NDRs (Figures 6D and 6E; Supplemental Table 6). Chromatin structure modulates critical processes such as gene transcription and DNA replication by controlling access to DNA, and regulatory factor binding sites frequently map to NDRs (Song et al., 2011; Zhang et al., 2012; Struhl and Segal, 2013). Over 95% of Arabidopsis S/MARs (clusters A, B, C, and E) overlap with at least one NDR (Figure 6D; Supplemental Table 6) and occur in regions of overall low nucleosome density (Figures 3A and 3B). The majority of these S/MARs are within 1 kb of a TSS, and matrix attachments in these regions could stabilize them in an open conformation. This could allow regulatory proteins to bind DNA, as was suggested previously for S/MARs associated with Arabidopsis TF genes (Tetko et al., 2006). Conversely, some S/MAR binding proteins appear to repress gene expression by occluding regulatory factor binding sites. The AGAMOUS gene, which has an S/MAR (Figure 4C) and multiple TF binding sites in its second intron (Sieburth and Meyerowitz, 1997; Ng et al., 2009; Dinh et al., 2012), may be repressed via such a mechanism. Similarly, the S/MAR at the TSS may help to repress SHORT ROOT expression in our cells (Figure 4F). In either case, S/MARs could provide an additional level of regulation that responds to specific developmental or environmental cues (Morisawa et al., 2000; Ng et al., 2009).
Although more than 95% of S/MAR clusters A, B, C, and E overlap with at least one NDR (Figure 6D; Supplemental Table 6), this only accounts for 16% of the 15,077 chr4 NDRs that we defined in this study. S/MAR mapping is limited by technical parameters and may emphasize those elements having the highest binding affinity. Thus, it is possible that more NDRs have the potential to act as S/MARs. However, our analysis of the sequence characteristics of S/MARs showed that they encompass a distinct subset of NDRs (Figure 6E; Supplemental Table 7).
Many DNA motifs previously shown to be potential matrix binding sites tend to be degenerate, AT rich, and contain poly(dA:dT) tracts (Figure 5). We broadened this observation by determining the 6-mer content of S/MARs and associated genomic regions. This revealed a striking enrichment of poly(dA:dT)-rich 6-mers in preference to AT-rich 6-mers in S/MARs (Figures 6A and 6B). This saw tooth pattern was unique to S/MARs and differed from the patterns seen with other closely associated sequences, including NDRs, promoters, introns, and TEs (Figure 6B). Poly(dA:dT) tracts have been shown to be functionally important for specific S/MARs (Adachi et al., 1989; Käs et al., 1989). Our analysis showing preferential enrichment of poly(dA:dT) tracts over other AT-rich DNA in a genome-scale data set suggested that poly(dA:dT) tracts are a general feature of S/MARs.
Poly(dA:dT) tracts have long been known to have important functions in eukaryotes (reviewed in Segal and Widom, 2009; Struhl and Segal, 2013) and are common in eukaryotic promoters (Iyer and Struhl, 1995). Even short poly(dA:dT) tracts disfavor nucleosome formation and are thought to increase the accessibility of DNA to TFs (Yuan et al., 2005; Field et al., 2008; Kaplan et al., 2009). Recent studies have shown that poly(dA:dT) tracts in promoters can fine-tune gene expression (Raveh-Sadka et al., 2012; Dadiani et al., 2013). In this context, it is striking that preferential enrichment for poly(dA:dT) tracts is a feature unique to S/MARs (Figures 6B and 6E).
S/MARs and poly(dA:dT) tracts have also been reported to enhance gene expression and block spreading of heterochromatin (Iyer and Struhl, 1995; Anderson and Widom, 2001; Bi et al., 2004; Field et al., 2008; Raveh-Sadka et al., 2012), thereby raising the question as to the relationship of S/MARs, poly(dA:dT) tracts, and NDRs. Both matrix preparations and nucleosomes display DNA binding preferences in vitro, and nucleosomes are more readily displaced from poly(dA:dT) tracts in vitro (Käs et al., 1989; Michalowski et al., 1999; Kaplan et al., 2009; Zhang et al., 2012). However, the relationship between poly(dA:dT) sequences and nucleosome binding in vivo may be more nuanced. While models based on the intrinsic binding of DNA sequences are good at estimating nucleosomal density (Kaplan et al., 2009), they do not account for the precise nucleosome positioning often observed in vivo. Instead, DNA binding proteins such as TFs, chromatin remodelers, and RNA polymerase II are thought to be critical for this process, especially for positioning the +1 nucleosomes of genes (Zhang et al., 2009b; Hughes et al., 2012). Intriguingly, cluster C S/MARs are ideally positioned to contribute to this process. Mechanistically, matrix proteins could prevent promiscuous binding of nucleosomes, possibly acting with proteins such as chromatin remodelers (Cai et al., 2003; Euskirchen et al., 2011). A productive area for future investigation would be to determine the extent to which S/MARs contribute to nucleosome positioning and thus facilitate the regulation of critical processes such as gene transcription and DNA replication.
METHODS
S/MAR Isolation
Arabidopsis thaliana Columbia-0 suspension cells line (Calikowski et al., 2003) were grown in Gamborg’s B5 basal medium with minor salt (Sigma-Aldrich 5893) supplemented with 1.1 mg/L 2,4-D, 3 mM MES, and 3% Suc and harvested from a 7-d split culture as previously described (Lee et al., 2010).
To prepare protoplasts for nuclei isolation the 7-d split culture was grown for 16 h, centrifuged (320g) for 10 min, and resuspended in 0.4 M mannitol, 10 mM MES cell wash buffer, pH 5.5, containing 1% cellulase (Onozuka RS; Research Products International), and 0.1% pectolyase (Onozuka Y23; Gold Biotechnology) and gently agitated for 1 h at 27°C.
Nuclei were isolated as described by Hall et al. (1991) with minor modifications. Arabidopsis protoplasts were suspended in 100 mL of nuclear isolation buffer (NIB), which contains 0.5 M hexylene glycol, 20 mM HEPES, pH 7.4, 20 mM KCl, 0.5 mM EDTA, 0.5% Triton X-100, 1% thiodiglycol, 50 μM spermine, 125 μM spermidine, 1 mM PMSF, and aprotinin (2 μg/mL) to lyse the plasma membrane to release the nuclei. Nuclei were filtered through 100-, 50-, and 30-μm nylon mesh to remove cellular debris and centrifuged at 4°C in NIB containing 15% Percoll at 600g for 10 min. The nuclear pellet was washed twice with NIB without Triton X-100, adjusted to 50% glycerol, and stored at −80°C for later use.
Nuclear halos were prepared as described (Hall et al., 1991) with minor modifications for Arabidopsis. One milliliter, containing 2 × 106 nuclei, was stabilized with 1 mM CuSO4 for 15 min at 37°C. Histones and other soluble proteins were removed with 2 mL of halo isolation buffer 2 (20 mM HEPES, pH 7.4, 2 mM EDTA, pH 7.4, 0.1% digitonin, 0.5 mM PMSF, aprotinin at 2 μg/mL, 10 μM E-64, and 100 mM lithium acetate) and 10 mM LIS (Supplemental Figure 1) and centrifuged and washed as described (Hall et al., 1991) for immediate use for S/MAR isolation or preparation of nuclear matrices.
Nuclear halos (4 × 106 nuclei) were digested for 3 h with EcoRI and HindIII. S/MARs and matrix proteins were isolated using centrifugation and digestion buffer washes as described (Hall et al., 1991). Arabidopsis S/MARs were isolated following an overnight proteinase digestion followed by extraction with phenol/chloroform/indole-3-acetic acid (Hall et al., 1991). The S/MAR-containing fraction was mixed with 4 μL of 150 μg mL−1 DNA GlycoBlue (Ambion), precipitated with NaOAc/ethanol, and resuspended in RT-PCR grade water (Ambion).
In Vitro Binding Assay
Putative S/MARs and non-S/MARs were fluorescently labeled using a three-primer method (Schuelke, 2000). The PCR mix for the fluorescent labeling contained 10 to 20 ng genomic template DNA, 2 mM MgCl2, 200 μM each deoxynucleotide triphosphate, 0.2 μM forward primer, which included an M13 -21 sequence tail (5′-TGTAAAACGACGGCCAGT-3′), 1 μM reverse primer, 0.05 μM fluorescently labeled M13 primer, Amplitaq 360 PCR reaction buffer, and one unit of Amplitaq DNA polymerase (Applied Biosystems). The M13 primer was 5′-labeled using a 700 nm near-infrared dye (LI-COR). Unique forward and reverse primers (Supplemental Data Set 10) were designed to flank the putative S/MAR region to produce PCR fragments of 0.3 to 1 kb. PCR was then used to produce fluorescently labeled fragments using the following conditions: initial denaturation at 94°C for 3 min; followed by 15 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C; followed by 25 cycles of 30 s at 94°C, 30 s at 50°C, and 60 s at 72°C; and a final for 7 min at 72°C. Labeled PCR fragments were purified using a QIAquick PCR purification kit (Qiagen) and resuspended in RT-PCR–grade water.
The labeled fragments were used for in vitro binding assays using matrices from either NT-1 tobacco (Nicotiana tabacum) or Arabidopsis nuclei (Hall et al., 1991; Michalowski et al., 1999). Nuclear halos were prepared and digested with 500 units of EcoRI and HindIII (Hall et al., 1991). The digested matrices were centrifuged and the pellet was washed twice with binding buffer (70 mM NaCl, 20 mM HEPES, pH 7.4, 20 mM KCl, 10 mM MgCl2, 0.1% digitonin, 1% thiodiglycol, 0.2 mM phenylmethylsulfonyl fluoride, aprotinin at 5 μg/mL, 10 μM E-64, and 1 mM phenantroline). In vitro binding was done for 3 h at 37°C in a mixture of 6 × 104 NT-1 or Arabidopsis nuclear matrices, 10 ng of end-labeled PCR fragments, and 10 μg of sonicated Escherichia coli competitor DNA. The mixture was then centrifuged 10 min (2000g, 4°C) to separate the S/MAR-containing pellet from the supernatant. The pellet was washed with digestion buffer and incubated in 50 μL lysis buffer for 16 h (37°C) as described above. Equal fractions (∼50%) of input DNA fragments (total), pellet, and supernatant fractions were separated on a TAE gel (1.5% agarose), and the bands were detected at 700 nm by infrared imaging (Odyssey; LI-COR).
Microarray Hybridization
Isolated S/MAR DNA fragments (target) and sonication-sheared Arabidopsis genomic DNA (reference) samples were subjected to whole-genome amplification using the GenomePlex WGA1 kit (Sigma-Aldrich) as described (O’Geen et al., 2006), omitting the initial random fragmentation step. Target and reference DNAs (10 ng) were linearly amplified for 14 cycles. The amplification product (10 ng) was then logarithmically amplified for 14 cycles and the amplified DNA was purified and concentrated to 200 to 250 ng/mL using a QIAquick PCR purification kit (Qiagen). Amplified target and reference samples (1.5 μg) were each labeled with Cy5 and Cy3 fluorescent dye–labeled 9-mer (TriLink Biotechnologies) and incubated 3 h (37°C) with 100 units (exo-) Klenow fragment (New England Biolabs) and deoxynucleotide triphosphate mix (10 mM each in Milli-Q water, pH 5.0; New England Biolabs). The reactions (100 μL) were terminated with 10 μL of 0.5 M EDTA, pH 8.0, precipitated with 11.5 μL of 5 M NaCl and an equal volume of isopropanol, and resuspended in water. Three labeling reactions produced 13 μg of both Cy5-labeled target and Cy3-labeled reference, which were mixed, evaporated, and resuspended in 39 μL of NimbleGen Hybridization Buffer (Roche). The target DNA was then cohybridized to a custom-designed NimbleGen tiling array for Arabidopsis chr4 (Roche NimbleGen), consisting of 174,978 isothermal probes in duplicate at a median tiling resolution of 100 bp. The hybridized arrays were washed using NimbleGen Wash Buffer System (Roche), dried by centrifugation, and coated with DyeSaver2 (Genisphere) diluted to 80%. Arrays were scanned on a GenePix 4000B scanner (Molecular Devices), and data were quantified using NimbleScan software (version 2.4.27).
Microarray Data Analysis
Microarray normalization and analysis was performed in R with Limma and Bioconductor (Gentleman et al., 2004; Smyth, 2005; R Development Core Team, 2009). Only probes specific for the euchromatic regions of chr4 were retained, and data were loess and quantile normalized using default settings. Duplicate probes were averaged, and a simple linear model was used to fit the biological replicates. The Pearson correlation for the probe ratios between the three biological replicates was >0.78, so the biological replicates were treated as technical replicates for subsequent analyses. Thus, the probe ratios are the average of 12 measurements: three biological replicates each with two hybridizations each and duplicate probes on each array.
The normalized and averaged probe ratios were exported as GFF files to the NimbleScan ChIP peak-finding function. Window sizes of 300, 400, 500, 600, and 700 bp were used, requiring three, four, five, five, or six positive probes, respectively. Peaks with an estimated FDR > 0.05 were removed, and the remaining peaks for all window sizes were merged to arrive at 1358 putative S/MARs. S/MARs were then mapped to the TAIR10 coordinate system.
Comparison of Experimental S/MARs with Predicted S/MARs
To determine the overlap between our S/MARs and the in silico predicted S/MARs (pS/MARs) for Arabidopsis (Rudd et al., 2004), the pS/MARs were mapped to the TAIR10 coordinate system (Swarbreck et al., 2008). S/MARs and pS/MARs were designated as overlapping unless separated by a gap of one base or more. All pS/MARs that mapped to the heterochromatic knob and pericentromeric DNA were removed from this analysis.
K-Means Clustering and Genomic Context S/MARs
To categorize S/MARs for subsequent analysis, k-means clustering was performed on S/MAR gene, exon, and TE content exploring the range of k = 2 to k = 25 with 20 maximum iterations and 1000 random starts (R Development Core Team, 2009) (Supplemental Figure 5 and Supplemental Table 3). Five clusters produced a good balance between biological relevance and variance within the clusters. S/MAR AT content was determined from the TAIR10 sequence and histograms used bins of two percentage points. S/MARs were further classified as either TSS- or TTS-proximal based on the distance from the S/MAR midpoint to the nearest gene end. For S/MARs that overlap genes, these distances were always determined relative to the overlapping gene. Histograms were calculated using 250-bp bins.
S/MARs and Chromatin Structure
Three data sources were used to analyze the chromatin structure of S/MARs and flanking sequences. A genome-wide predicted nucleosome occupancy is available for Arabidopsis TAIR8 (Kaplan et al., 2009; http://genie.weizmann.ac.il/software/nucleo_genomes.html). Nucleosome occupancy data for Arabidopsis shoots is available from next-generation sequencing of isolated mononucleosomes (Chodavarapu et al., 2010; accession number GSM543295). We defined NDRs from this data as those regions with mononucleosome sequence coverage in the bottom quartile, excluding regions with zero coverage as potential artifacts. Microarray results for chromatin immunoprecipitations to H3K4me2/1, H3K9me2, and H3K56ac as well as DNA 5mC for the Arabidopsis Columbia-0 cell line have been published (Tanurdzic et al., 2008). The microarray used for this analysis is lower resolution than our current platform, consisting of tiled PCR products with a mean length of ∼1 kb.
To analyze the nucleosome occupancy of S/MARs and flanking regions, S/MARs were aligned at their midpoint and windows were delineated extending 5 kb upstream and 5 kb downstream of the S/MAR midpoints. These windows were further subdivided into twenty 500-bp nonoverlapping bins. The mean nucleosome occupancy of the bins was then calculated from both the sequence predictions and next-generation sequence coverage and then averaged across each S/MAR cluster. For the histone modifications and DNA methylation, the intersection between regions bearing each modification and the 500-bp bins was determined and then averaged across each S/MAR cluster.
S/MARs and TEs
To determine the significance of the association between S/MARs and TEs, TEs were grouped by the 18 annotated superfamilies (Buisine et al., 2008), and the base pair overlap between the S/MARs and each TE-SF was calculated (Supplemental Table 4). If the association of S/MARs with the TE-SFs is random, then the expected TE-SF content of the S/MARs should be similar to the TE-SF content of chr4 (exclusive of the knob and pericentromere). Thus, the ratio of S/MAR TE-SF content to chr4 TE-SF content should be equal to one if the association is random. To test this null hypothesis, a permutation test was used to estimate a P value for the observed ratios. For the null data set, random start coordinates were chosen for 1358 regions, using the S/MAR lengths to assign end coordinates. The overlap between the random S/MARs and the TE-SFs was calculated, and this process was repeated 100,000 times to arrive at a null distribution for the ratios. P values were determined by the intersection of the observed ratio for the actual S/MARs with this null distribution, using the Bonferroni correction to adjust for multiple testing.
To determine the correlation between S/MAR enrichment in TE-SFs and the AT content of the TE-SFs, the mean AT content of the TE-SFs was calculated, and a linear model of the S/MAR TE-SF enrichment as a function of TE-SF AT content was determined.
Influence of S/MARs on Gene Expression
To examine the influence S/MARs on gene expression, genes that either overlap or are flanked by an S/MAR were classified by the distance from the gene TSS or TTS to the S/MAR midpoint and by the S/MAR cluster. Gene expression status was determined from an Affymetrix expression study of the Arabidopsis Columbia-0 cell line (Tanurdzic et al., 2008). Gene activity was defined as the percentage of genes in each class that were called as present, using MAS5 presence/absence calls (P ≤ 0.05) determined with Bioconductor (Gentleman et al., 2004). Statistical significance was determined by binomial tests in R with the null as the gene activity of chr4 genes, including those associated with an S/MAR.
Functional Clustering of Genes with a TSS Proximal S/MAR
GO enrichment analysis was performed with the Database for Annotation, Visualization and Integrated Discovery v 6.7 (DAVID) (Huang et al., 2009a, 2009b). As the background gene list, only chr4 genes in the regions under study were used. For TF genes, a gene was considered as encoding a TF only when annotated by both DAVID and the AtTFDB (Davuluri et al., 2003) as a TF gene.
DNA Motifs Associated with S/MARs
To determine if any previously identified DNA motifs are enriched in S/MARs, a list of 478 motifs was assembled that included 22 motifs believed to be specific for S/MARs as well as known plant cis-elements (Higo et al., 1999) (Supplemental Data Set 6). The occurrence of these motifs in chr4 and in the S/MARs was determined and the result was expressed as a motif density (counts per kilobase). To determine if the motifs were enriched in S/MARs, we used one-sample t tests with the motif density of chr4 as the null, using the Bonferroni correction to adjust the P values for multiple comparisons. Enrichment for the motifs was calculated by expressing the difference between the observed and expected density as a percentage of the expected density.
S/MARs 6-Mer Profiles
The occurrence of all possible 6-mers in the S/MARs and in chr4 was tabulated, and the results for each 6-mer and its reverse complement were combined. Enrichment for the 6-mers was calculated as above. P values for any observed enrichment were calculated from the binomial distribution using the occurrence of the 6-mer in chr4 as the null, adjusting the P values for multiple testing by the Bonferroni method. The 6-mers were also classified by both their AT content and poly(dA:dT) content and the enrichment for each class of 6-mer was then calculated.
Accession Number
Microarray data for S/MARs can be found at the Gene Expression Omnibus under accession number GSE45549.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Histones Removal from Arabidopsis Nuclei.
Supplemental Figure 2. In Vitro Nuclear Matrix Binding Assay Steps.
Supplemental Figure 3. Matrix Binding Competition between S/MARs and E. coli DNA.
Supplemental Figure 4. Modest S/MARs Enrichment in DNA Replication Initiation Zones and Early Replicating Regions.
Supplemental Figure 5. Selection of K for K-Means Clustering of S/MAR Data
Supplemental Figure 6. Relationship between TE-SF AT Content and Enrichment of TE-SF in S/MARs.
Supplemental Table 1. Distribution and Density of Genes, TEs, and S/MARs in Chr4 Regions.
Supplemental Table 2. Comparison of Mapped S/MARs and pS/MARs.
Supplemental Table 3. Summary of K-Means Clustering Results.
Supplemental Table 4. Data for TE Superfamily Enrichment Analysis.
Supplemental Table 5. Binomial Tests of Gene Activity.
Supplemental Table 6. NDR Content of S/MARs.
Supplemental Table 7. Comparison of S/MAR-Positive and S/MAR-Negative NDRs.
Supplemental Data Set 1. Summary Information for Arabidopsis Chr4 S/MARs.
Supplemental Data Set 2. Results of in Vitro Binding Assays.
Supplemental Data Set 3. Information for Genes Associated with an S/MAR.
Supplemental Data Set 4. Functional Cluster of Genes with TSS-Proximal S/MAR.
Supplemental Data Set 5. Information for All Chr4 Genes Related to Functional Cluster.
Supplemental Data Set 6. DNA Motifs Associated with S/MARs and Plant cis-Elements.
Supplemental Data Set 7. Motif Count for All S/MARs.
Supplemental Data Set 8. Statistics for DNA Motifs Enriched for DNA Motifs Enriched in at Least One S/MAR Cluster.
Supplemental Data Set 9. Statistics for all Nonredundant 6-Mer Pairs.
Supplemental Data Set 10. Primer Sequences to Amplify S/MAR Sequences and Negative Controls for in Vitro Binding.
Supplementary Material
Acknowledgments
We thank Iris Meier at Ohio State University for the Arabidopsis cell line and for discussions and advice. We thank present and former members of the replication group at North Carolina State University, especially Mariana Franco, Randall Shultz, Sharon Settlage, and our colleague Steven Spiker, for discussions and advice. We thank Robert Martienssen and members of his lab at Cold Spring Harbor Laboratory for suggestions on the project. This research was supported by National Science Foundation Grants DBI-0421651 and IOS-1025830.
AUTHOR CONTRIBUTIONS
M.A.F.-V. and P.E.P. performed the experiments. P.E.P., M.A.F.-V., and M.W.V. analyzed and evaluated the data. P.E.P., M.A.F.-V., B.S., L.H.B., M.W.V., W.F.T., and G.C.A. interpreted the data. W.F.T., L.H.B., and G.C.A. conceived the study and coordinated the research. P.E.P., M.A.F.-V., T.J.L., M.W.V., B.S., L.H.B., W.F.T., and G.C.A. wrote the article. All authors read and approved the final article.
Glossary
- LIS
lithium 3′-5′-diiodosalicylic acid
- S/MAR
scaffold/matrix attachment region
- NDR
nucleosome-depleted region
- chr4
chromosome 4
- FDR
false discovery rate
- TE
transposable element
- TSS
transcription start site
- TTS
transcription termination site
- TE-SF
TE superfamily
- GO
Gene Ontology
- TF
transcription factor
- NIB
nuclear isolation buffer
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adachi Y., Käs E., Laemmli U.K. (1989). Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 8: 3997–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrethsen J., Knol J.C., Jimenez C.R. (2009). Unravelling the nuclear matrix proteome. J. Proteomics 72: 71–81 [DOI] [PubMed] [Google Scholar]
- Alfonso-Parra C., Maggert K.A. (2010). Drosophila SAF-B links the nuclear matrix, chromosomes, and transcriptional activity. PLoS ONE 5: e10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.C., Hall G., Jr, Michalowski S., Newman W., Spiker S., Weissinger A.K., Thompson W.F. (1996). High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell 8: 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.C., Spiker S., Thompson W.F. (2000). Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol. Biol. 43: 361–376 [DOI] [PubMed] [Google Scholar]
- Anderson J.D., Widom J. (2001). Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 21: 3830–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova Z., SanMiguel P., Georgieva E., Bennetzen J.L. (1995). Matrix attachment regions and transcribed sequences within a long chromosomal continuum containing maize Adh1. Plant Cell 7: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova Z., Tikhonov A., Chen M., Bennetzen J.L. (1998). Matrix attachment regions and structural colinearity in the genomes of two grass species. Nucleic Acids Res. 26: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L. (2000). Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42: 251–269 [PubMed] [Google Scholar]
- Berezney R., Coffey D.S. (1974). Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun. 60: 1410–1417 [DOI] [PubMed] [Google Scholar]
- Bi X., Yu Q., Sandmeier J.J., Zou Y. (2004). Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24: 2118–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T. (1992). Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science 255: 195–197 [DOI] [PubMed] [Google Scholar]
- Boulikas T. (1993). Nature of DNA sequences at the attachment regions of genes to the nuclear matrix. J. Cell. Biochem. 52: 14–22 [DOI] [PubMed] [Google Scholar]
- Buisine N., Quesneville H., Colot V. (2008). Improved detection and annotation of transposable elements in sequenced genomes using multiple reference sequence sets. Genomics 91: 467–475 [DOI] [PubMed] [Google Scholar]
- Butaye K.M.J., Goderis I.J.W.M., Wouters P.F.J., Pues J.M.-T.G., Delauré S.L., Broekaert W.F., Depicker A., Cammue B.P.A., De Bolle M.F.C. (2004). Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39: 440–449 [DOI] [PubMed] [Google Scholar]
- Byrd K., Corces V.G. (2003). Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Han H.J., Kohwi-Shigematsu T. (2003). Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34: 42–51 [DOI] [PubMed] [Google Scholar]
- Cairns B.R. (2009). The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Calikowski T.T., Meulia T., Meier I. (2003). A proteomic study of the Arabidopsis nuclear matrix. J. Cell. Biochem. 90: 361–378 [DOI] [PubMed] [Google Scholar]
- Capco D.G., Wan K.M., Penman S. (1982). The nuclear matrix: Three-dimensional architecture and protein composition. Cell 29: 847–858 [DOI] [PubMed] [Google Scholar]
- Chodavarapu R.K., et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P.N., Garrard W.T. (1986). Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44: 273–282 [DOI] [PubMed] [Google Scholar]
- Dadiani M., van Dijk D., Segal B., Field Y., Ben-Artzi G., Raveh-Sadka T., Levo M., Kaplow I., Weinberger A., Segal E. (2013). Two DNA-encoded strategies for increasing expression with opposing effects on promoter dynamics and transcriptional noise. Genome Res. 23: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos M.K., Sieburth L.E. (2000). Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson L.A., Joh T., Kohwi Y., Kohwi-Shigematsu T. (1992). A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70: 631–645 [DOI] [PubMed] [Google Scholar]
- Dinh T.T., Girke T., Liu X., Yant L., Schmid M., Chen X. (2012). The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139: 1978–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G.M., Auerbach R.K., Davidov E., Gianoulis T.A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M.B., Snyder M. (2011). Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K., Ott S., Hansen A., Koentges G., Wernisch L. (2007). A comparative study of S/MAR prediction tools. BMC Bioinformatics 8: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y., Kaplan N., Fondufe-Mittendorf Y., Moore I.K., Sharon E., Lubling Y., Widom J., Segal E. (2008). Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLOS Comput. Biol. 4: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M., Frech K., Klingenhoff A., Cartharius K., Liebich I., Werner T. (2002). In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res. 12: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S., Matsunaga S., Yonemura M., Uchiyama S., Azuma T., Fukui K. (2004). Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol. Biol. 56: 225–239 [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Laemmli U.K. (1986). Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell 46: 521–530 [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T.I., Byrd K., Corces V.G. (2000). A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Hall G., Jr, Allen G.C., Loer D.S., Thompson W.F., Spiker S. (1991). Nuclear scaffolds and scaffold-attachment regions in higher plants. Proc. Natl. Acad. Sci. USA 88: 9320–9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9: 15–26 [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong R.L., Hamaguchi L., Busch M.A., Weigel D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009b). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell E., Liu W.M., Mei R. (2002). Robust estimators for expression analysis. Bioinformatics 18: 1585–1592 [DOI] [PubMed] [Google Scholar]
- Hughes A.L., Jin Y., Rando O.J., Struhl K. (2012). A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol. Cell 48: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V., Struhl K. (1995). Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenke B.H., Fetzer C.P., Stehle I.M., Jönsson F., Fackelmayer F.O., Conradt H., Bode J., Lipps H.J. (2002). An episomally replicating vector binds to the nuclear matrix protein SAF-A in vivo. EMBO Rep. 3: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I.K., Rogozin I.B., Glazko G.V., Koonin E.V. (2003). Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19: 68–72 [DOI] [PubMed] [Google Scholar]
- Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J., Segal E. (2009). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Izaurralde E., Laemmli U.K. (1989). Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J. Mol. Biol. 210: 587–599 [DOI] [PubMed] [Google Scholar]
- Keaton M.A., Taylor C.M., Layer R.M., Dutta A. (2011). Nuclear scaffold attachment sites within ENCODE regions associate with actively transcribed genes. PLoS ONE 6: e17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., Galande S. (2007). Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 9: 45–56 [DOI] [PubMed] [Google Scholar]
- Lam W.S., Yang X., Makaroff C.A. (2005). Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J. Cell Sci. 118: 3037–3048 [DOI] [PubMed] [Google Scholar]
- Lee T.-J., et al. (2010). Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet. 6: e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebich I., Bode J., Reuter I., Wingender E. (2002). Evaluation of sequence motifs found in scaffold/matrix-attached regions (S/MARs). Nucleic Acids Res. 30: 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann A.K., Platts A.E., Krawetz S.A. (2009). Differential nuclear scaffold/matrix attachment marks expressed genes. Hum. Mol. Genet. 18: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Luger K., Dechassa M.L., Tremethick D.J. (2012). New insights into nucleosome and chromatin structure: An ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner L.D., Hamlin J.L., Dijkwel P.A. (2003). The matrix attachment region in the Chinese hamster dihydrofolate reductase origin of replication may be required for local chromatid separation. Proc. Natl. Acad. Sci. USA 100: 3281–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski S.M., Allen G.C., Hall G.E., Jr, Thompson W.F., Spiker S. (1999). Characterization of randomly-obtained matrix attachment regions (MARs) from higher plants. Biochemistry 38: 12795–12804 [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Gasser S.M., Laemmli U.K. (1988). Scaffold attachment of DNA loops in metaphase chromosomes. J. Mol. Biol. 200: 101–109 [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M.-E., Laemmli U.K. (1984). Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell 39: 223–232 [DOI] [PubMed] [Google Scholar]
- Morisawa G., Han-Yama A., Moda I., Tamai A., Iwabuchi M., Meshi T. (2000). AHM1, a novel type of nuclear matrix-localized, MAR binding protein with a single AT hook and a J domain-homologous region. Plant Cell 12: 1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabirochkin S., Ossokina M., Heidmann T. (1998). A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 273: 2473–2479 [DOI] [PubMed] [Google Scholar]
- Ng K.-H., Ito T. (2010). Shedding light on the role of AT-hook/PPC domain protein in Arabidopsis thaliana. Plant Signal. Behavior 5: 200–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.H., Yu H., Ito T. (2009). AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 7: e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H., Nicolet C.M., Blahnik K., Green R., Farnham P.J. (2006). Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41: 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D., et al. (2008). Reconfiguration of genomic anchors upon transcriptional activation of the human major histocompatibility complex. Genome Res. 18: 1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniswamy S.K., James S., Sun H., Lamb R.S., Davuluri R.V., Grotewold E. (2006). AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J.R., Laemmli U.K. (1977). The structure of histone-depleted metaphase chromosomes. Cell 12: 817–828 [DOI] [PubMed] [Google Scholar]
- Raveh-Sadka T., Levo M., Shabi U., Shany B., Keren L., Lotan-Pompan M., Zeevi D., Sharon E., Weinberger A., Segal E. (2012). Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 44: 743–750 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2009). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B. (2009). The role of DNA shape in protein-DNA recognition. Nature 461: 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollini P., Namciu S.J., Marsden M.D., Fournier R.E.K. (1999). Identification and characterization of nuclear matrix-attachment regions in the human serpin gene cluster at 14q32.1. Nucleic Acids Res. 27: 3779–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd S., Frisch M., Grote K., Meyers B.C., Mayer K., Werner T. (2004). Genome-wide in silico mapping of scaffold/matrix attachment regions in Arabidopsis suggests correlation of intragenic scaffold/matrix attachment regions with gene expression. Plant Physiol. 135: 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18: 233–234 [DOI] [PubMed] [Google Scholar]
- Segal E., Widom J. (2009). Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 19: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L.E., Meyerowitz E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D.N., Wilson K.L. (2011). The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat. Rev. Mol. Cell Biol. 12: 695–708 [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2005). Limma Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R. Gentleman, V. Carey, S. Dut, R. Irizarry, and W. Huber, eds (New York Springer), pp. 397–420. [Google Scholar]
- Song L., et al. (2011). Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21: 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Segal E. (2013). Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 20: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Xie W., Xu F., Grunstein M., Li K.-C. (2009). Dissecting nucleosome free regions by a segmental semi-Markov model. PLoS ONE 4: e4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiki K., Kodama Y., Nakayama H., Shinmyo A. (2009). Determination of the in vivo distribution of nuclear matrix attachment regions using a polymerase chain reaction-based assay in Arabidopsis thaliana. J. Biosci. Bioeng. 108: 11–19 [DOI] [PubMed] [Google Scholar]
- Tanurdzic M., Vaughn M.W., Jiang H., Lee T.-J., Slotkin R.K., Sosinski B., Thompson W.F., Doerge R.W., Martienssen R.A. (2008). Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 6: 2880–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetko I.V., Haberer G., Rudd S., Meyers B., Mewes H.W., Mayer K.F. (2006). Spatiotemporal expression control correlates with intragenic scaffold matrix attachment regions (S/MARs) in Arabidopsis thaliana. PLOS Comput. Biol. 2: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, W.F., Spiker, S., and Allen, G.C. (2007). Matrix attachment regions and transcriptional gene silencing. In Annual Plant Reviews, Vol. 29: Regulation of Transcription in Plants, K. Gasser, ed (Hoboken, NJ: Wiley-Blackwell Publishing), pp. 136–161. [Google Scholar]
- Tikhonov A.P., Bennetzen J.L., Avramova Z.V. (2000). Structural domains and matrix attachment regions along colinear chromosomal segments of maize and sorghum. Plant Cell 12: 249–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drunen C.M., Oosterling R.W., Keultjes G.M., Weisbeek P.J., van Driel R., Smeekens S.C.M. (1997). Analysis of the chromatin domain organisation around the plastocyanin gene reveals an MAR-specific sequence element in Arabidopsis thaliana. Nucleic Acids Res. 25: 3904–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J.P., Dijkwel P.A., Mullenders L.H., Hamlin J.L. (1990). Replication forks are associated with the nuclear matrix. Nucleic Acids Res. 18: 1965–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kries J.P., Buhrmester H., Strätling W.H. (1991). A matrix/scaffold attachment region binding protein: Identification, purification, and mode of binding. Cell 64: 123–135 [DOI] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. (2005). Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang T., Wu Y., Jiang J. (2012). Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24: 2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. (2009a). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Moqtaderi Z., Rattner B.P., Euskirchen G., Snyder M., Kadonaga J.T., Liu X.S., Struhl K. (2009b). Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.