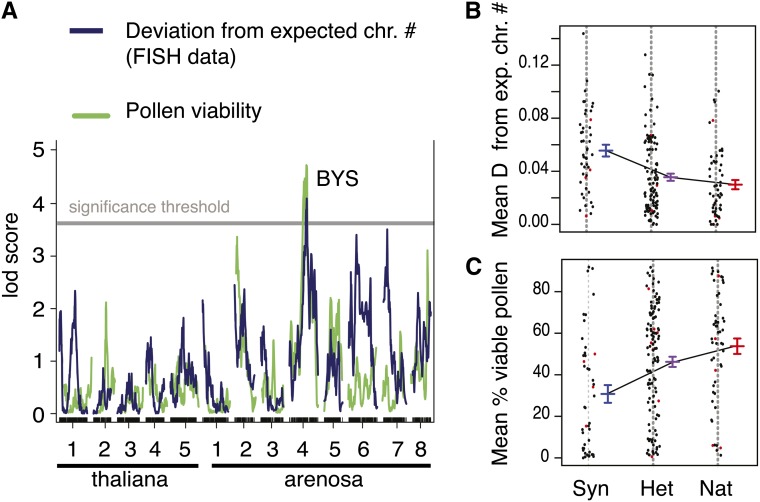
Figure 8.
Identification of a QTL Associated with Pollen Viability and Meiotic Irregularity.
(A) Results of QTL detection using standard interval mapping in R/qtl (see Methods for a detailed description of the phenotypes and the QTL mapping parameters used). LOD scores are plotted along the five A. thaliana and the eight A. arenosa chromosomes. Significance thresholds of 3.74 (deviation from expected chromosome number) and 3.73 (pollen viability) were determined based on 1000 permutations and correspond to P values < 0.05. The locus corresponding to the peak exhibiting significant LOD scores for both phenotypes was named BYS.
(B) and (C) Individual (cloud) and mean pollen viability (blue) (B) and individual (cloud) and mean deviation from expected chromosome number (meiotic irregularity in green) (C) for F2 plants that were homozygous for Syn (synthetic allopolyploid allele, left), heterozygous (middle), or homozygous for Nat (natural allopolyploid allele, right) at the BYS locus (indicated on [A] and located approximately at 16.9 Mb on scaffold 4 of the A. lyrata genomic reference). Standard errors are represented. Imputed genotypes are plotted in red.