During nitrogen starvation under mixotrophic conditions, thylakoids undergo extensive remodeling, leading to photosynthesis inactivation and increased chlororespiration. Degradation of specific photosynthetic protein complexes is triggered by the intracellular production of NO that originates from the rerouting of intracellular nitrite.
Abstract
Starving microalgae for nitrogen sources is commonly used as a biotechnological tool to boost storage of reduced carbon into starch granules or lipid droplets, but the accompanying changes in bioenergetics have been little studied so far. Here, we report that the selective depletion of Rubisco and cytochrome b6f complex that occurs when Chlamydomonas reinhardtii is starved for nitrogen in the presence of acetate and under normoxic conditions is accompanied by a marked increase in chlororespiratory enzymes, which converts the photosynthetic thylakoid membrane into an intracellular matrix for oxidative catabolism of reductants. Cytochrome b6f subunits and most proteins specifically involved in their biogenesis are selectively degraded, mainly by the FtsH and Clp chloroplast proteases. This regulated degradation pathway does not require light, active photosynthesis, or state transitions but is prevented when respiration is impaired or under phototrophic conditions. We provide genetic and pharmacological evidence that NO production from intracellular nitrite governs this degradation pathway: Addition of a NO scavenger and of two distinct NO producers decrease and increase, respectively, the rate of cytochrome b6f degradation; NO-sensitive fluorescence probes, visualized by confocal microscopy, demonstrate that nitrogen-starved cells produce NO only when the cytochrome b6f degradation pathway is activated.
INTRODUCTION
In most ecosystems, growth of plants and algae is restricted by nutrient availability: Nitrogen and phosphorus are often suboptimal in terrestrial and freshwater ecosystems, while primary production in oceans is limited by phosphorus near the coast and by iron in open oceans. Organisms cope with nutrient limitation by developing acclimation processes that combine general stress responses with changes in gene expression and metabolism that are specific to the limiting nutrient (Davies and Grossman, 1998; Merchant and Helmann, 2012), among which the induction of transporters to optimize the uptake of the limiting nutrient, of scavenging enzymes, and of recycling processes aimed at accessing alternative sources of this nutrient. As a paradigmatic example, cyanobacteria degrade their phycobilisomes to use them as nitrogen and carbon sources when facing the corresponding nutrient limitation (Yamanaka and Glazer, 1980; Collier and Grossman, 1994).
In Chlamydomonas reinhardtii, a unicellular green alga with great metabolic flexibility and for which there are powerful genetic tools (Grossman et al., 2007), acclimation to nutrient limitation has been extensively studied over the years (recently reviewed in Merchant and Helmann, 2012). C. reinhardtii expresses arylsulfatases (Lien and Schreiner, 1975; Schreiner et al., 1975), phosphatases (Quisel et al., 1996), and l-amino acid oxidase (LAO1; Vallon et al., 1993) when starved for sulfur, phosphorus, and nitrogen, respectively. It recycles ∼85% of its chloroplast sulfolipids upon sulfur limitation (Sugimoto et al., 2007, 2010) or part of its chloroplast DNA when starved for phosphorus or nitrogen (Sears et al., 1980; Yehudai-Resheff et al., 2007). As do other microorganisms facing nutrient shortage, C. reinhardtii undergoes cell cycle arrest and downregulation of photosynthesis when nutrient starved. Upon phosphorus and sulfur deprivation, decreased photosynthesis in C. reinhardtii has mainly been attributed to compromised photosystem II (PSII) activity (Wykoff et al., 1998), although this simple causal relationship has recently been revisited (Malnoë et al., 2014).
Nitrogen deprivation is of particular physiological significance for C. reinhardtii since, besides triggering acclimation to optimize nitrogen metabolism, it also induces ribosome remodeling and differentiation into sexually competent gametes (Siersma and Chiang, 1971; Martin et al., 1976; Bulté and Bennoun, 1990). Transcript profiling and proteomic studies have illustrated the extensive changes in gene expression undergone by nitrogen-starved C. reinhardtii (Miller et al., 2010; Boyle et al., 2012; Longworth et al., 2012), which reflect both the gamete differentiation program and the metabolic changes aimed at nitrogen recycling and carbon storage. This latter process has attracted increasing attention with the recent recognition of microalgae as a most promising source for renewable biofuel production (Wijffels and Barbosa, 2010). Macronutrient limitation is commonly used to divert C. reinhardtii metabolism toward biotechnologically valuable end products: Sulfur starvation triggers H2 photoproduction (Ghirardi et al., 2000), while nitrogen depletion boosts the storage of reduced carbon into lipid bodies (Hu et al., 2008). The latter consist mostly of neutral lipids, namely, triacylglycerol, the accumulation of which may further increase in the absence of starch biosynthesis (Wang et al., 2009; Work et al., 2010), even if the competition between these two pathways has been recently challenged (Siaut et al., 2011). Clearly, a better understanding of the changes in the bioenergetics of nitrogen-starved cells is required to get a refined picture of carbon allocation circuitries.
The photosynthetic properties of nitrogen-starved C. reinhardtii were studied more than two decades ago (Plumley and Schmidt, 1989; Peltier and Schmidt, 1991; Bulté and Wollman, 1992), at a time when knowledge of the thylakoid membrane protein complexity was still limited. A most noticeable feature of C. reinhardtii depleted in nitrogen sources in heterotrophic conditions under low light is photosynthetic downregulation due to a specific loss in cytochrome b6f complexes (Bulté and Wollman, 1992), rather than to PSII inactivation as reported for phosphorus and sulfur deprivation (Wykoff et al., 1998; Philipps et al., 2012). This loss in cytochrome b6f results from active proteolytic degradation (Bulté and Wollman, 1992; Majeran et al., 2000).
Here, we took advantage of the more recently acquired knowledge about the proteins that contribute (1) to the biogenesis and degradation of photosynthetic protein complexes and (2) to the redox poise of the photosynthetic membranes to investigate the extent of remodeling of thylakoids upon nitrogen starvation and to obtain insight as to which signals contribute to this acclimation process. This process converts these energy-producing membranes into a catabolic matrix that dissipates the energy stored in stromal reductants. It also leads to the unexpected loss of a number of cytochrome b6f biogenesis factors, which are subjected to the same conditional degradation as the cytochrome b6f complex itself. We demonstrate that these degradation processes are triggered by the intracellular production of NO and provide genetic evidence that NO originates from the rerouting of intracellular nitrite during nitrogen deprivation.
RESULTS
Upon Nitrogen Starvation, C. reinhardtii Becomes Photosynthetically Inactive by Losing Both the Cytochrome b6f Complex and Rubisco
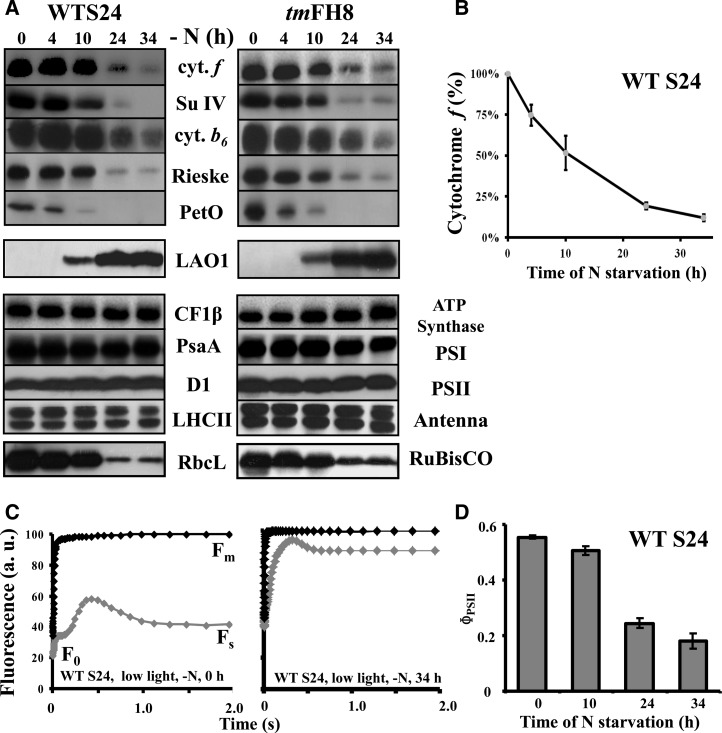
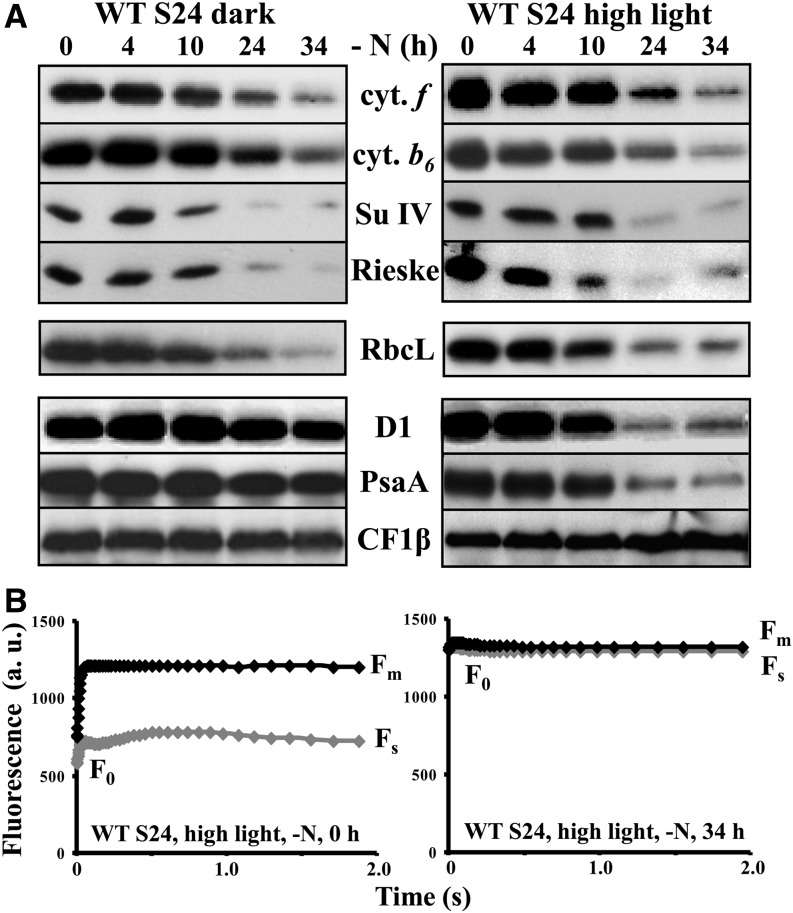
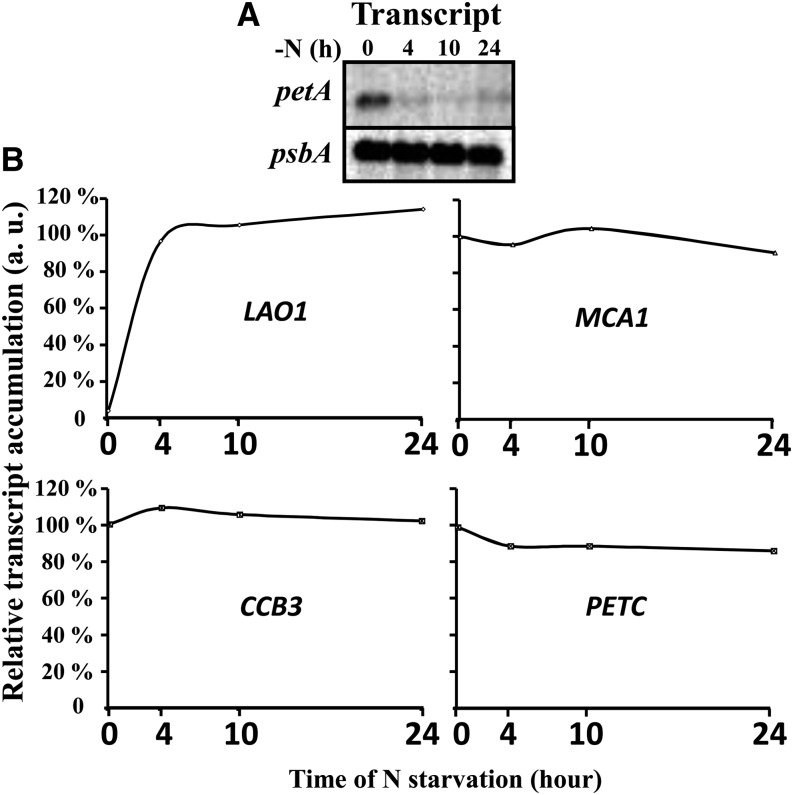
As previously reported (Bulté and Wollman, 1992), thylakoids specifically lose the cytochrome b6f complex upon nitrogen starvation of a wild-type strain of C. reinhardtii, derived from strain 137c (Harris, 2009), when grown heterotrophically (e.g., in the presence of acetate) in aerobic conditions under dim light (5 to 10 µE⋅m−2⋅s−1). In the typical experiment shown in Figure 1A for two strains of C. reinhardtii that have a wild-type phenotype for photosynthesis, WT-S24, and the wild-type-like strain tmFH8 (for details, see Table 1), all cytochrome b6f subunits, as well as its associated protein PETO (Hamel et al., 2000), dramatically decreased over time, as quantified in Figure 1B for cytochrome f. The loss of the cytochrome b6f complex, the major intersystem electron carrier that transfers electrons from PSII to photosystem I (PSI), leads to a major change in the quantum yield of PSII [ΦPSII= (Fm − Fs)/Fm, where F0, Fs, and Fm represent the initial, stationary, and maximal fluorescence level, respectively; Maxwell and Johnson, 2000; Figures 1C and 1D], as previously observed (Bulté and Wollman, 1992). In parallel, LAO1, a nitrogen scavenging enzyme that we previously identified as a marker of the cell response to nitrogen deprivation (Bulté and Wollman, 1992; Vallon et al., 1993), is induced (Figure 1A). Thus, nitrogen starvation triggers a cytochrome b6f–mediated downregulation of both linear and cyclic photosynthetic electron flows in C. reinhardtii. As previously observed (Majeran, 2002), Figure 1A also shows that the content in Rubisco decreases upon nitrogen starvation, with the same kinetics as that of the cytochrome b6f complex, further preventing photosynthetic reduction of carbon. By contrast, the accumulation of the other major photosynthetic protein complexes (LHCII, PSI, PSII, and ATP synthase) remained unaltered (Figure 1A). The selective loss of the cytochrome b6f complex and of Rubisco was not triggered by light since it was observed as well when nitrogen starvation was performed in complete darkness (Figure 2A, left panel). The kinetics of the loss remained similar whether starvation was performed in darkness or in low or high light (120 µE⋅m−2⋅s−1; Figure 2A). However, increasing the light intensity to 120 µE⋅m−2⋅s−1 generated additional photoinhibitory processes, targeting both PSI and PSII. This is shown by the loss of PsaA and D1 proteins (Figure 2A, right panel) and by the changes in the fluorescence patterns of wild-type cells deprived of nitrogen for 34 h under high light, which became similar with and without 3-(3,4-dichorophenyl)-1,1-dimethylurea (DCMU), no longer showing any variable fluorescence upon illumination and thus demonstrating the loss of PSII activity (Figure 2B).
Figure 1.
Cytochrome b6f Complexes and Rubisco Are Selectively Lost during Nitrogen Starvation under Low Light (5 to 10 µE⋅m−2⋅s−1).
(A) Whole-protein extracts of cells harvested at indicated time points (0, 4, 10, 24, and 34 h) after the onset of nitrogen starvation, probed with the antibodies indicated between the two panels. To detect all proteins investigated here but also in Figures 3A and 4, the samples were loaded several times on independent gels. WT-S24 (left panel) and strain tmFH8, which expresses tagged versions of MCA1 and TCA1 but is otherwise wild-type (right panel), are shown. Antibodies recognize the major cytochrome b6f subunits (top) or one major subunit of each other photosynthetic protein complex (bottom), whose abundance reflects that of the whole complex.
(B) Quantification of the loss of cytochrome b6f complex in strain WT-S24. Abundance of cytochrome f, expressed as a fraction of its initial amount, was quantified from phosphor imager scans of immunoblots revealed with 125I-protein A and normalized to the amount of the invariant CF1 subunit β to correct for variations in loading; mean of three independent experiments ± sd.
(C) Kinetics of fluorescence induction of dark-adapted WT-S24 cells at the onset (left panel) and at the end (34 h; right panel) of the nitrogen starvation, recorded in the presence (gray curves) or in the absence (black curves) of DCMU (5 µM final). Relevant parameters (Fm, Fs, and F0) are shown. Curves were normalized to the maximum fluorescence recorded in the presence of DCMU. a.u., arbitrary units.
(D) Change in photosynthetic efficiency of WT-S24 cells during nitrogen starvation assessed by the quantum yield of PSII [ΦPSII = (Fm − Fs)/Fm]; mean of four independent experiments ± sd.
Table 1. Mutant Strains Used in This Work.
| Strain | Genotype | Phenotype | Ref. |
|---|---|---|---|
| WT 4γ | NIT1 NIT2 mt+a | Wild-type for photosynthesis and nitrogen assimilation; grows on nitrate and nitrite | [1] |
| nit1-137− | nit1-137 mt−a | Lacks NaR; cannot grow on nitrate, but grows on nitrite | [1] |
| nit2-124 | nit2-124, mt+a | Lacks expression of NaR, NiR, and HANiT; cannot grow on nitrate nor nitrite | [1] |
| WT S24 | nit1-137 nit2-124 mt−; results of multiple back-crosses of 137c mt+ and mt− strains | Our reference strain | [1] |
| Lacks expression of NiR, NaR, and HANiT; cannot grow on nitrate nor nitrite | [1] | ||
| 21gr | ΝΙΤ1 ΝΙΤ2 mt+ | Wild-type for photosynthesis and nitrogen assimilation | CC-1690 |
| M3 | Δ(NII1 NRT2;2 NRT2;1 NAR2 NIT1) ::NIT1 ::(NRT2;2 NRT2;1) ::NAR2 | Lacks NiR; cannot grow on nitrate nor nitrite | [2] |
| M4 | Δ(NII1 NRT2;2 NRT2;1 NAR2 NIT1) ::NIT1 | Lacks NiR and HANiT; cannot grow on nitrate nor nitrite | [2] |
| nit4-104 | nit4-104 NIT1 NIT2 mt+ | Lacks the MoCo cofactor of NaR and XOR; cannot grow on hypoxanthine, nitrate, or nitrite | [3] CC-2900 |
| Tft5 | nit1-305 ::NIT1 mt+ | Complemented NaR mutant | [4] |
| dum22 | nit1-137 nit2-124 mt− [Δcob, Δnd4] | Lacks mitochondrial cytochrome bc1 and NADH complexes. | [5] |
| nda2-RNAi | nda2RNAi nit1-137 nit2-124 | Knockdown of the NDA2 gene | [6] |
| ptox2 | ptox2::aphVIII nit1-137 nit2-124 | Knockout the PTOX2 gene | [7] |
| {ΔpetB} | nit1-137 nit2-124 mt+ {ΔpetB} | Cytochrome b6f mutant deleted for the petB gene | [8] |
| ptox2 {ΔpetB} | ptox2::aphVIII nit1-137 nit2-124 {ΔpetB} | The ptox2 mutant carrying a deletion of the petB gene | [7] |
| tmFH8 | mca1-6 ::MCA1-HA tca1-8 ::TCA1-Fl nit1-137 nit2-124 mt- | Expresses tagged versions of MCA1 and TCA1; otherwise wild-type for photosynthesis | [9] |
| mH ftsh1-1.2+ | mca1-6 ::MCA1-HA ftsh1-1 nit1-137 nit2-124 | Generated by cross mH mt− × ftsh1-1 mt+ | [1] |
| mH {clpP-AUU} | mca1-6 ::MCA1-HA nit1-137 nit2-124 {clpP-AUU} | Generated by cross mH mt− × {clpP-AUU} mt+ | [1] |
By convention, chloroplast or mitochondrial genotypes, when relevant, follow the nuclear genotype and are written between brackets and braces, respectively, while parentheses indicate gene clusters, either deleted or inserted in the strain. CC numbers refer to strains obtained from the Chlamydomonas Resource Center (http://chlamycollection.org). References: [1] this work; [2] Navarro et al. (2000); [3] Chamizo-Ampudia et al. (2013); [4]; Kindle et al. (1989); [5] Remacle et al. (2006); [7] Houille-Vernes et al. (2011); [8] Kuras and Wollman (1994); [9] Boulouis et al. (2011).
Generated from the cross WT-S24 mt− × 21gr mt+ (CC-1690)
Figure 2.
Loss of the Cytochrome b6f Complex Does Not Depend on the Incident Light.
(A) Whole-cell protein extracts from WT-S24 deprived of nitrogen under total darkness (left panel) or high light (120 µE⋅m−2⋅s−1, right panel), analyzed as in Figure 1A.
(B) Kinetics of fluorescence induction of dark-adapted (30 min) WT-S24 cells, recorded at the 0- and 34 h-time points of nitrogen starvation in high light. The Fm, Fs, and F0 parameters are shown. a.u., arbitrary units.
Nitrogen Starvation Induces Overexpression of the Chlororespiratory Enzymes
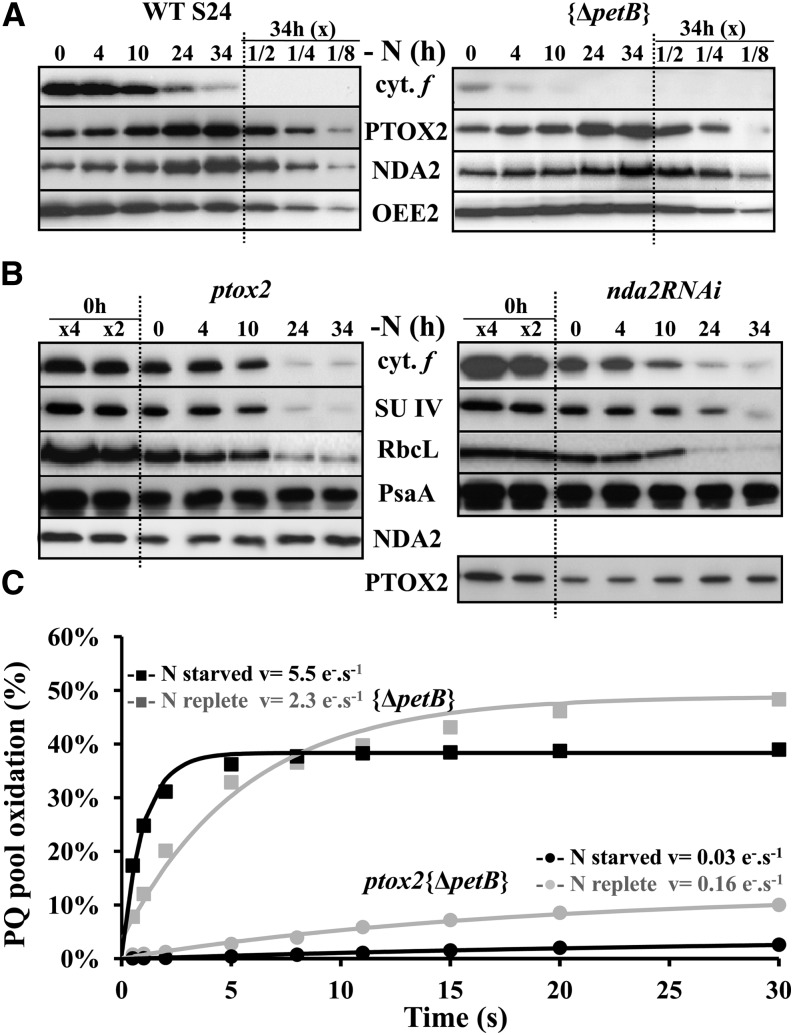
The loss of cytochrome b6f complexes during nitrogen starvation hampers the light-induced reoxidation of the plastoquinone (PQ) pool. However, in darkness or dim light (5 to 10 µE⋅m−2⋅s−1), the redox status of the PQ pool is primarily determined by chlororespiration (Bennoun, 1982). This prompted us to monitor two major chlororespiratory enzymes: the chloroplast-localized type-II NAD(P)H dehydrogenase NDA2 (Desplats et al., 2009) and PTOX2 (for Plastid Terminal Oxidase, isoform 2), the major oxidase involved in chlororespiration (Houille-Vernes et al., 2011). Both behaved opposite to the subunits of the cytochrome b6f complex: They increased about 4-fold over 34 h of nitrogen starvation under dim light (Figure 3A, left panel). This increase was independent of the light regime, still occurring when nitrogen starvation was performed in darkness or at 120 µE⋅m−2⋅s−1 (Supplemental Figure 1). The respective increases in PTOX2 and NDA2 proved largely independent from one another: NDA2 still increased about 3-fold in a ptox2 knockout mutant starved for nitrogen (Houille-Vernes et al., 2011), while PTOX2 increased 2-fold in a knockdown nda2RNAi strain (Jans et al., 2008) (Figure 3B). These two strains lost the cytochrome b6f complexes with similar kinetics as in the wild type (Figure 3B). Conversely, a mutant lacking the cytochrome b6f complex still showed a 4-fold increase in NDA2 and PTOX2 ({ΔpetB}, Figure 3A, right panel). Thus, the loss of cytochrome b6f complexes and the upregulation of these two enzymes are independently triggered during nitrogen starvation.
Figure 3.
Nitrogen Starvation Leads to Increased Chlororespiration.
(A) Steady state accumulation of the chlororespiratory enzymes PTOX2 and NDA2, probed with specific antibodies in WT-S24 (same samples as in Figure 1A) and {ΔpetB} strains subjected to nitrogen depletion for the indicated time points. A dilution series of the samples starved for 34 h is shown for the ease of quantification. Accumulation of the PSII subunit OEE2 provides a loading control.
(B) Abundance of PTOX2 or NDA2 in strains defective for the expression of the other enzyme, nda2RNAi or ptox2, respectively. The accumulation of diagnostic cytochrome b6f subunits was assessed in the same samples as that of the PSI subunit PsaA (as a loading control). A dilution series of the initial sample is shown for the ease of the comparison.
(C) Percentage of oxidation of the PQ pool in a cytochrome b6f–defective mutant, {ΔpetB}, or in a double mutant lacking both the cytochrome b6f complex and the terminal oxidase PTOX2, ptox2 {ΔpetB}, in nitrogen-replete (gray curves) and nitrogen-starved (34 h; black curves) conditions. The PQ pool was first fully reduced by a saturating flash, and the number of electron acceptors available for PSII was measured as a function of the duration of the subsequent dark period. The initial slope of the curve provides a measure of the rate of oxidation of the PQ pool.
To investigate the functional consequences of the increased content in chlororespiratory enzymes upon nitrogen starvation, we compared, as described by Bennoun (2001), the rate of reoxidation of the PQ pool in nitrogen-replete conditions or after 34 h of nitrogen starvation. This measurement is best performed in a cytochrome b6f mutant that still shows increased accumulation of chlororespiratory enzymes upon nitrogen starvation (Figure 3A). Cells, preilluminated for 2 s to fully reduce the PQ pool, were returned to darkness for a variable time and the extent of plastoquinol reoxidation was then measured as the area above the fluorescence induction curve in an ensuing illumination. The PQ reoxidation rate increased 2.4-fold after 34 h of nitrogen starvation (Figure 3C). In a ptox2 {ΔpetB} double mutant, lacking both PTOX2 and the cytochrome b6f complex, the rate of reoxidation of the PQ pool decreased 5-fold after 34 h of nitrogen starvation (Figure 3C), in agreement with the increase in NDA2 during nitrogen starvation that was still observed in the ptox2 mutant strain (Figure 3B).
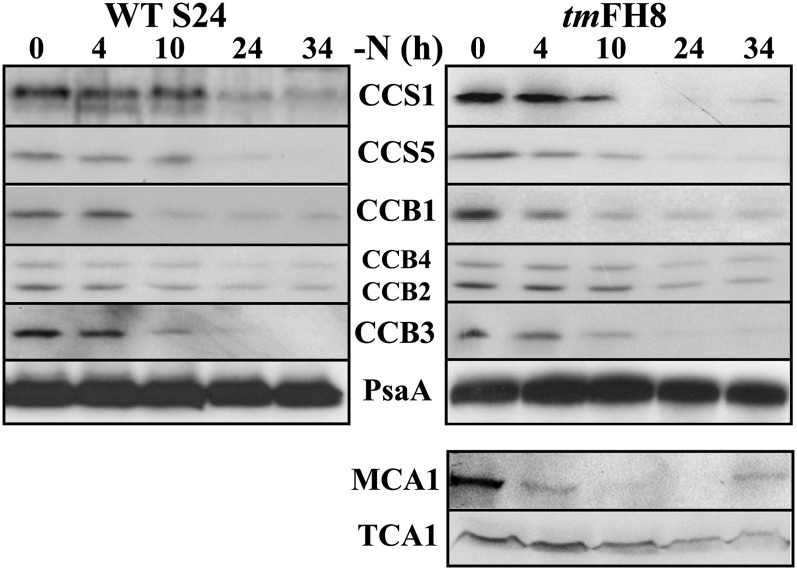
Upon Nitrogen Starvation, Most Proteins Specifically Required for the Biogenesis of Cytochrome b6f Complexes Are Lost
Many proteins, hereafter referred to as cytochrome b6f biogenesis factors, are specifically required for the biogenesis of the cytochrome b6f complex. These include proteins involved in the apo- to holoconversion of c-type cytochromes f and b6, respectively, the CCS (for C-type cytochrome synthesis; Xie et al., 1998) and CCB (for cofactor binding on cytochrome b6f subunit PetB; Kuras et al., 2007) factors, as well as trans-acting factors governing the expression of one of its chloroplast-encoded subunits, such as MCA1 and TCA1 that govern the expression of the chloroplast petA gene (maturation/stability and translation, respectively, of the cytochrome b6f subunit encoded by the petA gene; Wostrikoff et al., 2001; Loiselay et al., 2008). To detect the latter two, we resorted to the tmFH8 strain that expresses epitope-tagged versions of MCA1 and TCA1 (Raynaud et al., 2007; Boulouis et al., 2011). The tmFH8 strain, when starved of nitrogen under dim light, lost cytochrome b6f complexes with the same kinetics as WT-S24 (Figure 1A). Notably, the various cytochrome b6f biogenesis factors decreased significantly upon nitrogen starvation in both strains, as shown in Figure 4. MCA1 was even lost earlier than the subunits of the cytochrome b6f, which is consistent with its short half-life in N-replete conditions (Raynaud et al., 2007), but we note that its steady state accumulation increased slightly again toward the end of the experiment. CCS1, CCS5, and CCB1 disappeared with the same kinetics as cytochrome b6f subunits, while TCA1, CCB2, and CCB4 showed a more limited loss compared with other cytochrome b6f biogenesis factors. Thus, when deprived of nitrogen sources, C. reinhardtii specifically eliminates most of the proteins required for cytochrome b6f activity, whether structural subunits of the protein complex or biogenesis factors.
Figure 4.
Cytochrome b6f Biogenesis Factors Are Lost during Nitrogen Starvation.
Accumulation of cytochrome b6f biogenesis factors, assessed in the same samples as in Figure 1A, using specific antibodies directed against those proteins that are indicated between the two panels. Accumulation of MCA1 and TCA1 was probed using antibodies directed against the tags (HA for MCA1 and Flag for TCA1).
The Loss of the Cytochrome b6f Biogenesis Factors Does Not Result from a Transcriptional Shutdown but from Their Increased Proteolytic Degradation, Largely Mediated by the FtsH Protease
Most cytochrome b6f biogenesis factors being encoded by nuclear genes, as is the Rieske subunit (PETC), we investigated whether their loss could originate from a decreased transcription, as a part of the extensive transcriptional changes that develop during nitrogen starvation (Miller et al., 2010; Toepel et al., 2011). The expression of LAO1 was, as expected (Miller et al., 2010), rapidly induced in nitrogen-starved cells, while the chloroplast petA mRNA disappeared (Figure 5A) due to the loss of its stabilizing factor MCA1 (Figure 4). By contrast, the level of PETC, CCB3, and MCA1 transcripts remained invariant during the first 24 h of nitrogen starvation (Figure 5B), although the level of their protein products already strongly decreased (Figures 1A and 3). Our results on the accumulation of the PETC mRNA during nitrogen starvation differ from those of Miller et al. (2010), which may be accounted for by differences in time points (24 versus 48 h), in the light regime and in culture conditions. Thus, the decrease in these cytochrome b6f complex biogenesis factors cannot be attributed to an inhibition of gene transcription in the nucleus but rather to some posttranscriptional mechanisms.
Figure 5.
Transcriptional Downregulation Is Not the Cause of the Loss of the Nucleus-Encoded Cytochrome b6f Subunits and Biogenesis Factors.
(A) Steady state accumulation of the petA mRNA during nitrogen starvation, probed by RNA gel blotting. Accumulation of the psbA mRNA is shown as a loading control.
(B) Variations in transcript abundance, normalized to the initial value for MCA1, CCB3, and PETC genes, assessed by quantitative RT-PCR. Accumulation of the LAO1 transcript was normalized to the amount reached after 4 h of starvation. a.u., arbitrary units.
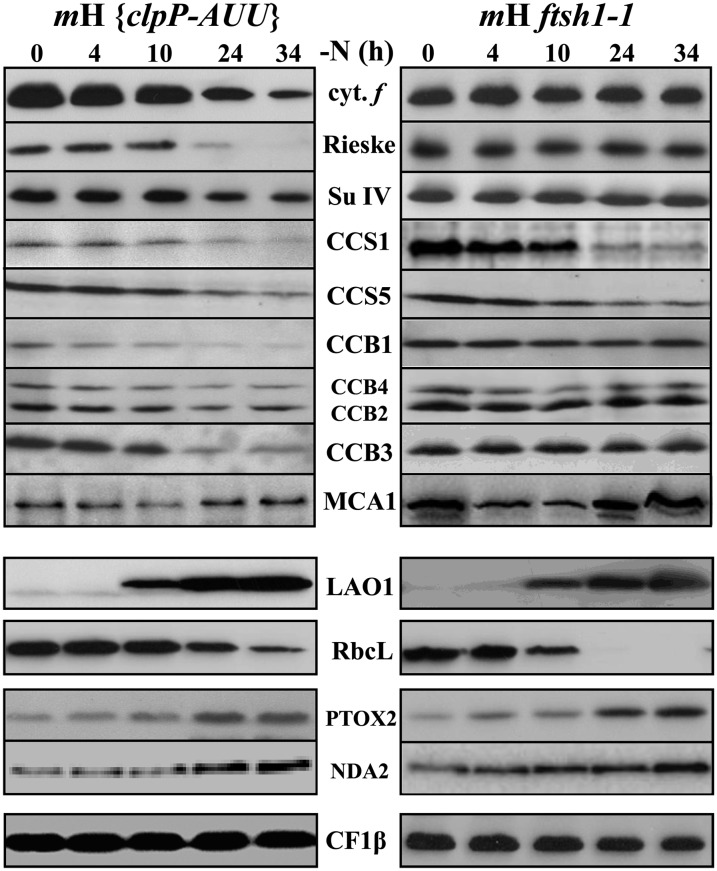
We have previously shown that the loss of cytochrome b6f subunits in nitrogen-starved C. reinhardtii cells was a posttranslational event, partially mediated by the Clp protease, as it was delayed in the clpP1-AUU mutant that displays a 4-fold reduced abundance of the catalytic ClpP subunit (Majeran et al., 2000). Here, we revisited these observations, now extended to the whole set of cytochrome b6f biogenesis factors and to Rubisco. We also studied the ftsh1-1 mutant defective for FtsH activity because of an R420C substitution altering an Arg finger essential for its ATPase and protease activities (Karata et al., 1999). Both ftsh1-1 and clpP1-AUU mutations were placed in a background expressing tagged MCA1 by sexual crossing. The loss of cytochrome b6f subunits was delayed in the mH {clpP-AUU} mutant, in agreement with our previous study (Majeran et al., 2000), and fully prevented in the mH ftsh1-1 mutant, as shown in Figure 6 (compared with WT-S24 in Figure 1A). CCB proteins behaved as the cytochrome b6f subunits, showing delayed degradation in the clpP-AUU mutant and complete preservation in the ftsh1-1 mutant. MCA1 was sensitive to both proteases, being markedly stabilized by both mutations, so that its increase at a late stage of nutrient stress was more visible than in the wild type. By contrast, the soluble stromal protein Rubisco was insensitive to the inactivation of the membrane-embedded protease FtsH, while its loss was somewhat delayed in the mutant showing attenuated expression of the soluble protease Clp. The loss of CCS1 and CCS5 proved poorly sensitive to the altered activity of either Clp or FtsH proteases, as expected from the topology of these proteins that are little exposed to the stromal side of the thylakoid membranes where both proteases are active.
Figure 6.
The Loss of Cytochrome b6f Complex Subunits and Biogenesis Factors Results from Their Increased Proteolytic Degradation.
The fate of cytochrome b6f subunits and biogenesis factors, LAO1, Rubisco, and PTOX2, was assessed as described in Figure 1 by immunoblots using the antibodies against those proteins listed between the two panels in two strains both expressing the tagged version of MCA1 and either defective for FtsH activity (mH ftsh1-1; right panel) or showing a 4-fold reduced accumulation of the Clp protease (mH {ClpP-AUU}; left panel). The β-subunit of the chloroplast ATP synthase is shown as a loading control.
Together, these results highlight the critical role of proteolysis in the disappearance of cytochrome b6f subunits and biogenesis factors, which independently are targeted for degradation by multiple proteases upon nitrogen starvation.
The Degradations of Cytochrome b6f Subunits, Cytochrome b6f Biogenesis Factors, and Rubisco Are Triggered by the Same Signals
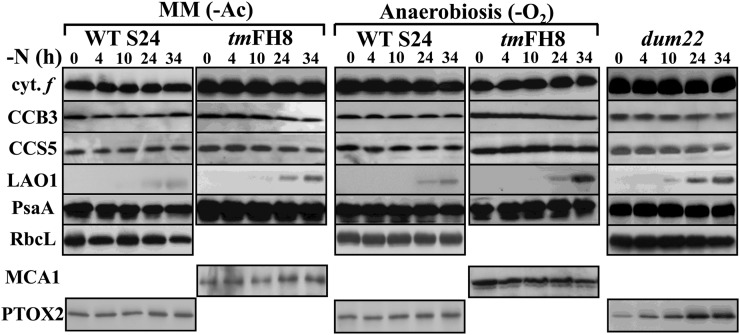
The loss of cytochrome b6f complex subunits is a regulated process that is no longer observed when C. reinhardtii cells are starved of nitrogen in the absence of acetate as a reduced carbon source or when mitochondrial respiration is impaired (Bulté and Wollman, 1992). To understand whether the losses of cytochrome b6f biogenesis factors and of Rubisco were similarly regulated, we resorted to three distinct experimental conditions. First, we grew C. reinhardtii WT-S24 and tmFH8 strains in phototrophic conditions (i.e., in minimum medium), before subjecting them to nitrogen starvation in the absence of acetate. Alternatively, we prevented mitochondrial respiration by placing WT-S24 and tmFH8 strains in anaerobic conditions during nitrogen starvation in the presence of acetate. Finally, we deprived of nitrogen the dum22 mutant, which is defective for mitochondrial respiration because of a deletion of the cob gene and a partial deletion of the nd4 gene (Remacle et al., 2006). As shown on Figure 7, neither Rubisco nor cytochrome f, taken here as a typical cytochrome b6f complex subunit, nor the cytochrome b6f biogenesis factors, exemplified by CCB3, CCS5, and MCA1, decreased significantly upon nitrogen starvation in these three experimental conditions. Induction of LAO1 could still be detected in all cases but was delayed and much weaker than when nitrogen starvation was imposed in the presence of acetate and in aerobic conditions, which suggests that the two phenomena are under common control. Thus, when ATP production primarily depends on photosynthesis, C. reinhardtii preserves the cytochrome b6f subunits and biogenesis factors as well as Rubisco, even in the absence of nitrogen sources, showing that all these proteins are under common regulation. The induction of PTOX2 was extremely limited upon nitrogen starvation in the absence of acetate or in anaerobiosis but was not affected in the respiratory mutant dum22 that was starved of nitrogen in aerobic and heterotrophic conditions, suggesting that the lack of mitochondrial respiration per se is not sufficient to prevent the induction of chlororespiration.
Figure 7.
Cytochrome b6f Subunits and Biogenesis Factors Are Preserved When Mitochondrial Respiration Is Impaired.
The accumulation of cytochrome b6f subunits and biogenesis factors, LAO1, Rubisco, and PTOX2, was studied as in Figure 1A in WT-S24 and tmFH8 strains deprived of nitrogen sources under low light, either in the absence of reduced carbon in the medium (-Ac; left panel) or in anaerobic conditions (-O2; cultured with gentle shaking, 50 rpm, in sealed Erlenmeyer flasks; middle panel), or in the respiratory mutant dum22 grown under 80 µE⋅m−2⋅s−1 (right panel). The antibodies used are directed against those proteins indicated on the left of the figure, and the accumulation of PsaA provides a loading control.
In an attempt to identify a physiological signal triggering these degradation processes, we explored the possibility that changes in protein phosphorylation associated with state transitions played a critical role once nitrogen starvation had started. This possibility was ruled out using various mutant strains blocked either in State I or in State II that all showed similar losses in cytochrome b6f subunits and biogenesis factors (Supplemental Figure 2). We also examined a cascade hypothesis whereby the loss of one protein would trigger the loss of the other proteins upon nitrogen starvation. The best candidates for priming this degradation cascade were those that were lost first, MCA1, CCS5, or CCB3. However, mca1-6, ccs5-T78, or ccb3-1 mutants still lost the remaining cytochrome b6f biogenesis factors (Supplemental Figure 3) as did the other mutants specifically lacking only one of the nitrogen-sensitive cytochrome b6f biogenesis factors. Neither the presence of either of the major photosynthesis proteins nor the activity of the cytochrome b6f complex itself at the beginning of nitrogen starvation was a prerequisite for these losses, as shown in Supplemental Figure 4.
Testing the Role of NO in the Loss of the Cytochrome b6f Complex
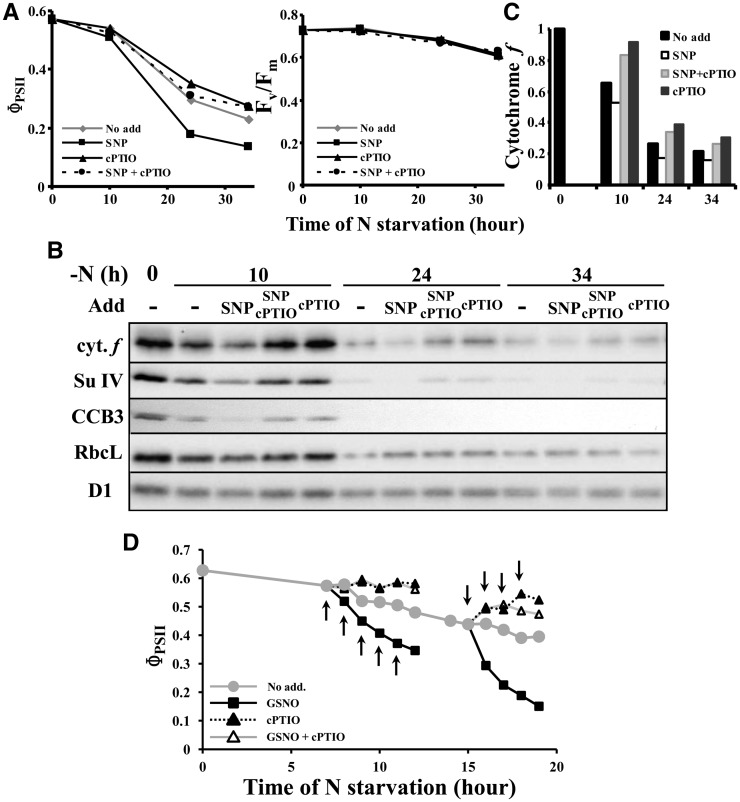
We then wondered which posttranslational modification might independently target the various cytochrome b6f subunits and biogenesis factors for proteolytic degradation. Nitrosylations frequently occur on metalloproteins, such as heme binding proteins. They represented a reasonable candidate to target cytochrome b6f subunits and biogenesis factors for degradation (see Discussion). To assess the possible involvement of NO in this process, we attempted to modify its concentration during nitrogen starvation and looked for possible changes in the kinetics of photosynthesis inactivation and loss in cytochrome b6f subunits, as studied by fluorescence and immunoblotting. We first used sodium nitroprusside (SNP) as a NO producer and 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) as a NO scavenger molecule. Since SNP slowly releases NO over a long time range (up to 10 h according to Ederli et al. [2009] and Mur et al. [2011]), both molecules were added 1 h after the onset of nitrogen starvation. Compared with the untreated control, ΦPSII decreased faster in cells treated with 1 mM SNP and more slowly in cells treated with 0.1 mM cPTIO (Figure 8A). The contrast between the two treatments was even larger in cells treated three times with SNP or cPTIO 1, 3, and 6 h after the beginning of nitrogen starvation (Supplemental Figures 5A and 5B). These changes in ΦPSII values corresponded to actual changes in the rate of degradation of cytochrome b6f and biogenesis factors, as shown in Figure 8B: At a given time point, the level of cytochrome f, Subunit IV, or CCB3 level was lower and higher in SNP- or cPTIO-treated cells than in the control cultures (see quantifications for cytochrome f in Figure 8C). PSII was not affected by SNP and/or cPTIO addition, as judged from the maximum PSII quantum yield [Fv/Fm = (Fm − F0)/Fm] and the D1 content (Figures 8A and 8B; Supplemental Figure 5A). We also note that Rubisco degradation showed some sensitivity to the addition of SNP and cPTIO at 10 h of nitrogen starvation but that their effect was no longer significant at later time points. That the faster degradation of the cytochrome b6f complex in the presence of SNP should be attributed to the release of NO rather than to other effects of the SNP molecule is supported by the antagonistic effect of cPTIO: When SNP and cPTIO were added simultaneously, the loss in cytochrome b6f, instead of being faster, was even delayed when compared with the untreated control (Figure 8).
Figure 8.
The Degradation of Cytochrome b6f Complex Is Accelerated in the Presence of NO.
(A) and (B) Strain WT-S24 was transferred to nitrogen-free medium and supplemented 1 h later with water (no add), 1 mM SNP, 0.1 mM cPTIO, or 1 mM SNP plus 0.1 mM cPTIO. Samples, harvested at 10, 24, and 34 h, were assessed for changes in photosynthetic activity (ΦPSII; [A]) and for the accumulation of cytochrome b6f subunits, CCB3, and Rubisco (B). In (A), the maximum quantum yield of PSII Fv/Fm shows there is no NO-mediated inactivation of the PSII.
(C) Quantification of cytochrome f accumulation in the typical experiment shown in (B).
(D) Strain WT-S24 was transferred to nitrogen-free medium and supplemented at the time points indicated by arrows with water (no add), 0.1 mM GSNO, 0.1 mM cPTIO, or 0.1 mM GSNO plus 0.1 mM cPTIO. Aliquots of the cultures were harvested every hour for the measure of the photosynthetic activity (ΦPSII).
To further confirm that the effect of SNP was indeed due to the release of NO, we used S-nitrosoglutathione (GSNO) as an alternative NO donor. Compared with SNP, GSNO induces a much faster burst of NO within the first hour after its addition to a culture (Ederli et al., 2009; Mur et al., 2011), allowing a better temporal control of NO production. GSNO (0.1 mM) was therefore added to the starvation medium, either before any significant cytochrome b6f degradation or when this process had already started (i.e., 7 and 15 h after the onset of nitrogen depletion, respectively). GSNO addition was then repeated every hour. When compared with an untreated control culture, addition of GSNO led in both cases to a much faster decrease of ΦPSII, already visible after the first addition of GSNO (Figure 8D). We noted that, in some experiments, the maximum quantum yield of PSII decreased slightly after the third addition of GSNO, suggesting that at this stage, PSII also became affected. When other aliquots of the same cultures were treated with cPTIO (0.1 mM), at the same time point as GSNO, the decrease in ΦPSII observed during the nitrogen starvation was fully prevented (cells depleted of nitrogen for 7 h) or even reversed (cells depleted of nitrogen for 15 h). This suggests that the cytochrome b6f complex is first reversibly inactivated before being degraded. The simultaneous addition of both drugs fully reversed the action of GSNO (Figure 8D), demonstrating that its effect was indeed due to the release of NO.
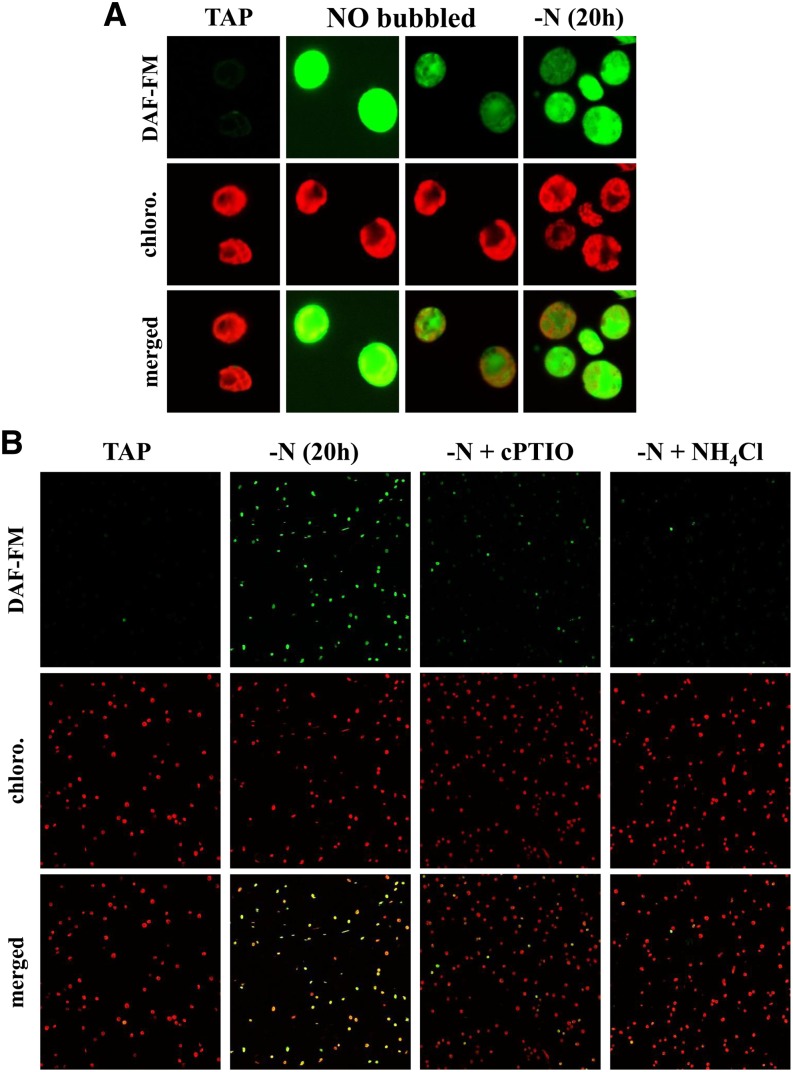
C. reinhardtii Cells Starved for Nitrogen Produce NO
To detect in situ endogenous NO production, we examined C. reinhardtii cells by confocal microscopy, after 1 h incubation with the NO-specific fluorescent probe 4-amino-5-methylamino-2',7'-difluoro-fluorescein diacetate (DAF-FM DA). This permeant and nonfluorescent molecule enters the cell where it is esterified into the nonpermeant and weakly fluorescent DAF-FM molecule. In the presence of NO (more specifically of its oxidation products N2O3 or NO+), it is converted into the highly fluorescent DAF-FM triazol derivative (Xie and Shen, 2012).
Figure 9A shows images of WT-S24 cells, examined for their chlorophyll autofluorescence (recorded between 647 and 797 nm, referred to as “red” signal) and DAF-FM T fluorescence (493 to 599 nm, referred to as the “green” signal). In cells kept in nitrogen-replete (Tris-acetate-phosphate [TAP]) medium, the chloroplast was easily distinguished by its chlorophyll autofluorescence. In these cells, a very weak green autofluorescence signal colocalized with the red signal (Figure 9A, left panels) and was also observed in the absence of DAF-FM DA (Supplemental Figure 6). When TAP-grown cultures were bubbled for 3 min with 90% N2/10% NO, a strong green fluorescence signal was readily detected (Figure 9A) that was not restricted to the chloroplast compartment. The weaker green signal within the chloroplast may result either from a lower accumulation of NO and/or DAF DA in this compartment or, more likely, from the reabsorption of the green fluorescence by photosynthetic pigments. Strikingly, most WT-S24 cells starved for nitrogen sources during 20 h also displayed a strong green fluorescence signal (Figures 9A, right panel, 9B, and 10A) that we attribute to the presence of NO, as the signal was largely decreased when cPTIO (0.1 mM) was added to the starvation medium half an hour before the addition of DAF-FM DA (Figure 9B).
Figure 9.
The DAF-FM Signal Specifically Reflects the Accumulation of NO.
Visualization of in vivo NO production by nitrogen-starved C. reinhardtii cells using confocal microscopy. Green and red fluorescence were due to the NO indicator DAF-FM and chlorophylls, respectively.
(A) Fluorescence pattern of isolated WT-S24 cells grown in TAP medium (TAP), grown in TAP medium and bubbled for 3 min with 10% NO (NO bubbled), or deprived of nitrogen for 20 h [-N(20 h)]. For a better view of the distribution of the green fluorescence in NO-bubbled cells, the same picture is shown on the right, with a lower gain during the acquisition of the green signal.
(B) Confocal images of cell populations grown in TAP (TAP) or deprived of nitrogen in the presence of acetate for 20 h (-N). Imaging was also performed on the same starved culture supplemented with either 0.1 mM cPTIO or 0.5 mM NH4Cl.
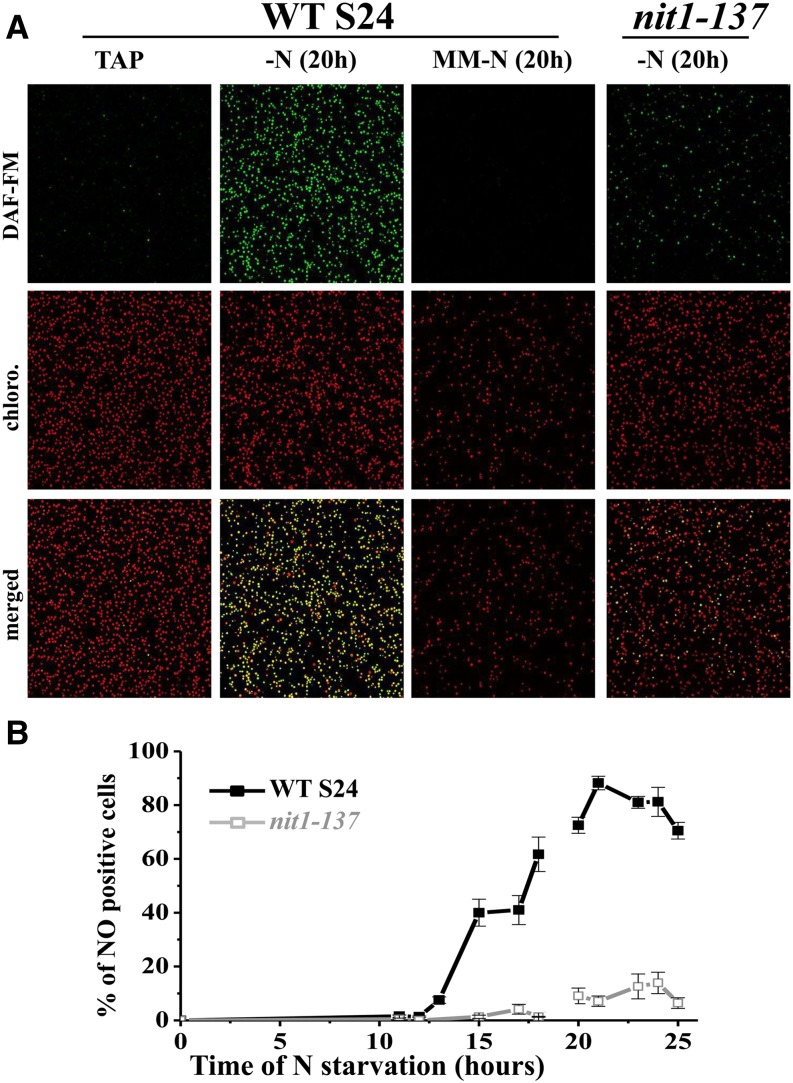
Figure 10.
Upon Nitrogen Starvation, C. reinhardtii Cells Produce NO.
(A) Confocal images of cell populations from the WT-S24 strain grown in TAP (TAP), deprived of nitrogen in the presence of acetate for 20 h (-N), or deprived of nitrogen in the absence of acetate (MM-N) or from the nit1-137 strain deprived of nitrogen in the presence of acetate for 20 h.
(B) Percentage of NO-positive cells over the time course of nitrogen starvation for the WT-S24 and the nit1-137 strains. For practical reasons, as indicated by the discontinuity of the plot, the first (0 to 18 h) and the second (20 to 25 h) parts represent independent starvation experiments. Error bars reflect, for each time point, the fraction of cells for which the DAF-FM fluorescence level is in between that of NO-negative and NO-positive cells.
To get a statistical view of NO production over a large population, we conducted another starvation experiment in which cells were concentrated 20 times before confocal imaging performed over larger area (5 × 5 tiles). WT-S24 cultures grown in TAP, or cultures deprived of nitrogen for <12 h, almost completely lacked NO positive green fluorescent cells (Figure 10). The percentage of NO-positive cells then increased progressively with the duration of nitrogen starvation, reaching its maximum value of ∼90% of the cells after 19 to 21 h of starvation, before decreasing slowly in the next hours (Figure 10B). To further test the relationships between nitrogen starvation, NO evolution, and the degradation of the cytochrome b6f complex, the same experiment was performed on a nitrogen-depleted culture of the nit1-137 mutant strain that lacks nitrate reductase (NaR) and does not lose the cytochrome b6f subunits and biogenesis factors (see below). That strain showed little detectable accumulation of NO over the time course of the experiment, with <15% of the cells exhibiting green fluorescence after 20 h of starvation (Figures 10A and 10B). In addition, when strain WT-S24 was subjected to nitrogen depletion in the absence of acetate (minimal medium), a condition that prevents the loss of the cytochrome b6f complex, almost no NO positive cells were observed after 20 h of starvation (Figure 10A) (i.e., at the time when the accumulation of NO is maximal in cultures starved of nitrogen in the presence of acetate). Last, the addition of 0.5 mM NH4Cl together with the DAF-FM DA probe in a culture deprived of nitrogen for 19 h led to the disappearance of green fluorescence 1 h later (Figure 9B).
Nitrite Accumulation Increases Cytochrome b6f Complex Degradation
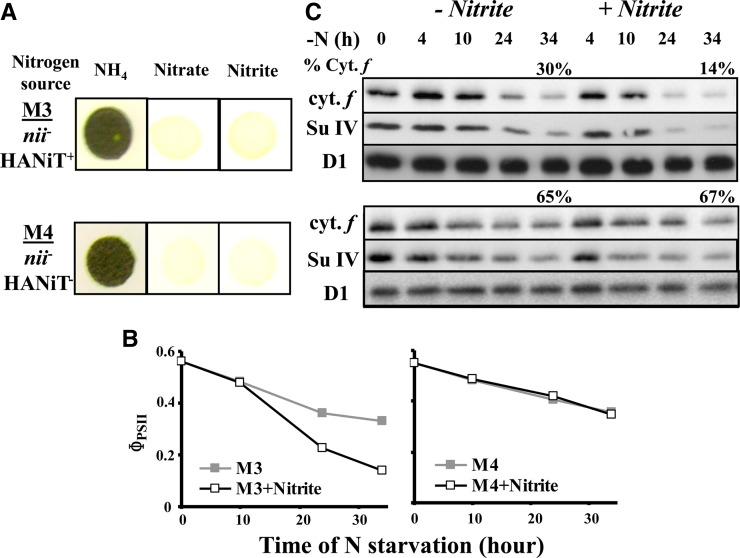
In an attempt to identify the source of NO produced upon nitrogen starvation, we investigated the possible contribution of nitrite (NO2−), a major source of NO through its reduction by a variety of pathways (reviewed in Gupta et al., 2011). Because nitrite reductase (NiR), by reducing nitrite to ammonium, should limit its availability for NO production, we compared the rate of degradation of cytochrome b6f complexes in mutants defective in NiR, when starved in the absence or presence of exogenously added nitrite. We compared the behavior of strains M3, carrying a deletion of the structural NII1 gene encoding NiR and M4, which, in addition, lacks the NTR2;2, NTR2;1, and NAR2 genes encoding components of the high-affinity nitrate/nitrite transporter HAN(i)T (Navarro et al., 2000; reviewed in Galvan and Fernández, 2001). As expected, strains M3 and M4 were unable to grow on nitrite or nitrate as their sole nitrogen source (Figure 11A). They lost the cytochrome b6f complex during nitrogen starvation, as revealed by their ΦPSII (Figure 11B) and cytochrome f and subunit IV contents (Figure 11C), albeit to a lower extent and with delayed kinetics compared with our reference strain WT-S24. When the nitrogen-free medium was supplemented with 0.1 mM NaNO2 1, 3, and 5 h after starvation had started, the loss of the cytochrome b6f complex was accelerated in strain M3 (Figures 11B and 11C). By contrast, strain M4 proved insensitive to the addition of nitrite, in agreement with its lack of import ability (Figures 11B and 11C). In these strains, as in WT-S24, PSII activity (measured by Fv/Fm) remained unchanged during nitrogen starvation, whether nitrite was added or not. This result supports the view that NO, efficiently produced from nitrite (Sakihama et al., 2002), accelerates the loss of the cytochrome b6f complex.
Figure 11.
The Degradation of the Cytochrome b6f Complex Is Accelerated in the Presence of Nitrite.
(A) Growth characteristics of the M3 and M4 strains on solid TAP medium (NH4) and on solid nitrogen-free medium supplemented with 2 mM KNO3 (Nitrate) or 2 mM NaNO2 (Nitrite).
(B) and (C) Strains were resuspended in nitrogen-free medium, and 0.1 mM NaNO2 was added to the starvation medium 1, 4, and 6 h after the onset of the starvation. Aliquots of the culture were harvested at the indicated time point and assessed as above for photosynthetic electron transfer (B) and accumulation of cytochrome f, subunit IV, and the PSII subunit D1 (C) as loading control. The percentage of cytochrome f remaining at the end of starvation is indicated.
Genetic Evidence for a Critical Role of Nitrite in the Loss of Cytochrome b6f Complexes
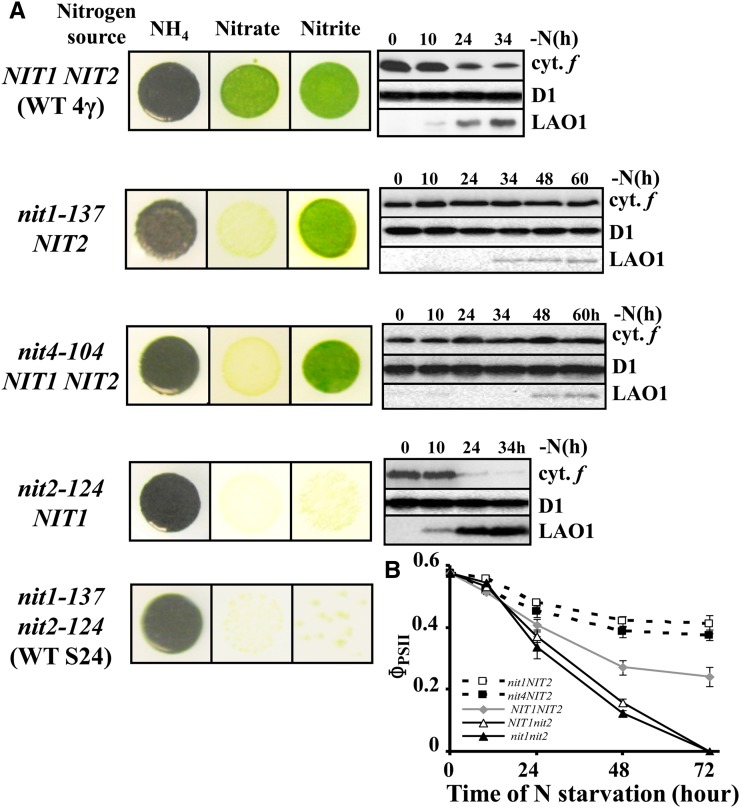
In a second series of experiments, we examined how the metabolism of nitrite in strains differing in their nitrate assimilation pathway correlated with the loss of the cytochrome b6f complex. We compared three groups of strains that were either wild type for nitrate/nitrite assimilation, defective for only nitrate assimilation, or defective for nitrate and nitrite assimilation (Figure 12A).
Figure 12.
The Kinetics and Extent of Cytochrome b6f Complex Degradation Depends on the Genotype of the Strain with Regard to Nitrogen Metabolism.
(A) Growth characteristics and accumulation of cytochrome f and LAO1 during nitrogen starvation of strains WT 4γ (NIT1 NIT2), nit1-137, nit4-104, nit2-124, and WT-S24 (nit1-137 nit2-124). Accumulation of the PSII subunit D1 is shown as a loading control. For nit1-137 NIT2 and nit4-104 NIT2 strains, the starvation was performed over 60 h to assess the stability of the cytochrome b6f complex and the delayed and limited induction of LAO1.
(B) Change in photosynthetic electron transfer (ΦPSII) in the same strains starved for nitrogen sources.
Our reference strain WT-S24 and all of the photosynthesis mutants used in this and previous studies to characterize the loss of cytochrome b6f complexes upon nitrogen starvation are derived from wild-type 137c. Like their 137c ancestor, they carry two mutations in genes required for nitrogen assimilation: nit1-137, a deleterious substitution that changes the axial ligand H542 of heme b5 to Q in the NIT1 structural gene encoding NaR, and nit2-124, an insertion of the TOC1 transposon into the first exon of the NIT2 gene (N. Tourasse, Y. Choquet, and O. Vallon, unpublished results), encoding the major nitrate/nitrite regulator (González-Ballester et al., 2004; Camargo et al., 2007). The NIT2 gene product is required for the expression of the NIT cluster that includes the gene for NaR, NIT1, together with genes for nitrate/nitrite assimilation (NII1, NTR2;1, NTR2;2, and NAR2) (Quesada et al., 1993, 1998; reviewed in Fernandez and Galvan, 2008). Therefore, a nit2-124 strain can grow neither on nitrite nor on nitrate as a nitrogen source, as is the case for a nit1-137 nit2-124 strain, exemplified here by WT-S24 (Figure 12A). These strains rapidly lose the cytochrome b6f complex upon nitrogen starvation (as shown in Figures 1A, 1D, 12A, and 12B). Strains that are wild-type for nitrogen metabolism (i.e., growing on either nitrate or nitrite as a nitrogen source) (Figure 12A) also lose the cytochrome b6f complex but to a lower extent and with delayed kinetics, as exemplified by wild-type 4γ in Figures 12A and 12B. We conclude that in a nit2 genetic background, (1) the absence of NiR, a potent sink for nitrite, allows enhanced accumulation of nitrite that feeds the production of NO, resulting in the rapid degradation of the cytochrome b6f complex; and (2) NaR is dispensable for NO production in the absence of NiR.
We then examined strains nit1-137 and nit4-104, which can grow on nitrite but not on nitrate because they lack only an active NaR, being respectively mutated in the NIT1 structural gene or in the CNX1E gene whose product is required for the biogenesis of the Molybdopterin cofactor (MoCo) (Chamizo-Ampudia et al., 2013). In contrast with the strains above, these two NaR-defective strains did not lose cytochrome b6f complexes upon nitrogen starvation, as shown by the preserved abundance of cytochrome f, even over a longer time scale (Figure 11A). Accordingly, they showed a very limited decrease in ΦPSII (Figure 12B). Similar results were obtained with other NaR-deficient strains, listed in Supplemental Table 1. Moreover, complementation of the nit1-305 mutant by the wild-type NIT1 gene (strain Tft5 in Kindle et al., 1989) restored active degradation of cytochrome b6f complexes (Supplemental Figure 7). The nit1-137 (Figures 10A and 10B) and nit4-104 strains (data not shown) also produce very little NO when examined by confocal microscopy. Thus, three types of behavior can be observed: (1) no loss of cytochrome b6f in NaR-deficient nit1, nit4, or nit7 mutants, (2) delayed loss in strains possessing both NaR and NiR, and (3) rapid loss in nit2 mutants lacking the whole nitrate/nitrite uptake and reduction pathways. In genetic crosses, these phenotypes consistently cosegregated with the corresponding genotypes, as detailed in Supplemental Tables 1 and 2. We conclude that the activity of NaR, although dispensable for NO production in the absence of NiR, is required when this nitrite sink is active. In the latter case, it helps keep the level of nitrite, formed at the expense of nitrate, high enough to sustain NO production.
DISCUSSION
Nitrogen Starvation Remodels Thylakoid Membranes into a Matrix for Catabolic Processes
When deprived of nitrogen, C. reinhardtii kept in mixotrophic conditions (in the presence of acetate) undergoes growth arrest after two rounds of cell division (Siersma and Chiang, 1971; Martin and Goodenough, 1975) while losing its ability to fix carbon. This occurs through two coregulated processes: the selective depletion of Rubisco, the key enzyme of the Benson-Calvin cycle, a phenomenon that is also observed in sulfur-starved cultures of C. reinhardtii (Zhang et al., 2002), and the degradation of an essential component of the photosynthetic electron transfer chain, the cytochrome b6f complex, required both for NADPH production through photosynthetic linear electron flow and ATP production through cyclic and linear electron flows. Thus, C. reinhardtii cells adapt their metabolism to nitrogen shortage by inhibiting their ability to reduce carbon. This regulation, ultimately aimed at maintaining balanced availability of nitrogen and carbon resources, does not result from an overflow/imbalance in the photoproduction of NADPH or ATP, as it is independent of the light regime, of the photosynthetic activity, and of state transitions. It is counteracted only by a block in mitochondrial respiration or by a drop in the intracellular flux of reduced carbon when acetate is omitted from the starvation medium, two conditions where the viability of C. reinhardtii rests exclusively on photosynthesis. Similarly, iron-limited C. reinhardtii cells maintain their photosynthetic apparatus when kept in phototrophic conditions, whereas photosynthesis decreases and respiration is prioritized in cells grown in the presence of acetate in the medium (Terauchi et al., 2010). Remarkably, C. reinhardtii has developed a conditional regulation of its photosynthetic properties to preserve its viability.
During nitrogen starvation in mixotrophic conditions, the bioenergetic contribution of the thylakoid membranes changes dramatically since the decreased photosynthesis comes along with a marked increase in chlororespiration. The latter change was suggested about two decades ago on functional and molecular bases (Peltier and Schmidt, 1991), the latter of which turned out to be erroneous since mitochondrial cytochromes (Atteia et al., 1992) were mistaken for chlororespiration components. Here, we demonstrate that, in parallel with the block in photosynthetic electron transfer due to the loss of cytochrome b6f complexes, the actual chlororespiratory enzymes NDA2 and PTOX2 markedly increased, leading to increased electron flux through chlororespiration, as we measured here by fluorescence techniques. However, the increase in chlororespiration is not mechanistically triggered by the degradation of the cytochrome b6f complex since it was still observed in mutants lacking this complex or in respiratory mutants that do not lose the cytochrome b6f complex. The increased chlororespiration has two major functional consequences. First, it contributes to counteract acetate assimilation and to limit starch/lipid storage by stimulating a chloroplast oxidative pathway. From the point of view of biofuel production, tuning down chlororespiration may be worth exploring in the future. The increase in PTOX2 also provides a safety valve, protecting PSII from low-light photoinhibition by plugging additional electron acceptors after the PQ pool. Oxygen thus becomes a genuine electron sink at low light in cytochrome b6f–defective conditions (i.e., after 10 to 20 h of nitrogen deprivation). Yet, the chlororespiratory pathway is not efficient enough to prevent PSI and PSII photodestruction at higher light intensities, as documented in this study.
Cytochrome b6f Biogenesis Factors: A Protein Network Specifically Sensitive to the Nitrogen Status
In earlier studies (Xie et al., 1998; Wostrikoff et al., 2001; Kuras et al., 2007; Raynaud et al., 2007; Loiselay et al., 2008), we identified several proteins that participate exclusively in the biogenesis of cytochrome b6f complexes. Here, we show that most of them behaved as did the cytochrome b6f subunits: They are degraded upon nitrogen starvation, and this degradation process is blocked when nitrogen starved cells cannot perform mitochondrial respiration or are kept in photoautrophic conditions. This concerted proteolysis is under the control of the FtsH protease with some contribution of the Clp protease and of a so far unidentified protease active on the lumenal side of the thylakoid membrane where the transmembrane CCS1 and CCS5 proteins protrude. That cytochrome b6f or the CCB proteins behave as genuine substrates for the FtsH protease upon nitrogen starvation is consistent with its role in the degradation of misassembled cytochrome b6f complexes in nitrogen-replete conditions and in the degradation of CCB4 in the absence of CCB2 (Malnoë et al., 2014). In our attempts to identify a mechanism responsible for the degradation of such a diverse array of soluble and integral proteins, we were able to exclude a cascade hypothesis whereby the loss of one protein would trigger the degradation of the other cytochrome b6f subunits or biogenesis factors, as was shown for the increased degradation of the various subunits of a photosynthetic protein complex in mutants lacking only one of its subunits (Wollman et al., 1999; Choquet and Vallon, 2000). As an alternative, we looked for posttranslational modification that would target these proteins for degradation.
NO Is Involved in the Remodeling of Photosynthesis during Nitrogen Starvation
The increase in PTOX2 upon nitrogen starvation was reminiscent of the behavior of the homologous alternative oxidase in mitochondria, which is triggered by NO-induced inhibition of cytochrome oxidase (Huang et al., 2002). In many biological systems, nitrosylation participates in signaling (reviewed in Astier et al., 2011, 2012) or induces protein degradation (Souza et al., 2000, Kim et al., 2004; Lee et al., 2008; Y. Wang et al., 2010; Wei et al., 2011; Tang et al., 2012; Jaba et al., 2013). Here, we reasoned that most of the cytochrome b6f subunits and biogenesis factors belong to heme binding complexes or participate in the biogenesis of heme binding proteins, which are particularly prone to nitrosylation (Thomas et al., 2003; Angelo et al., 2008). Indeed, in a recent proteomic study, several cytochrome b6f subunits were found to be nitrosylated in C. reinhardtii (Morisse et al., 2013).
In agreement with this hypothesis, confocal microscopy with an NO-sensitive fluorescence probe allowed us to detect NO production, but only in C. reinhardtii cells losing the cytochrome b6f complex (i.e., starved for nitrogen in normoxic conditions and in the presence of acetate). By contrast, C. reinhardtii deprived of nitrogen in phototrophic or anoxic conditions or with a genetic background that did not lead to cytochrome b6f degradation produced little, if any, NO upon nitrogen starvation. Furthermore, treatments increasing NO levels, such as addition of NO donors or nitrite in the starvation medium, increased the rate and extent of cytochrome b6f complex degradation, whereas NO scavengers had the opposite effect.
Intracellular Nitrite Is the Most Likely Source of NO Produced during Nitrogen Starvation
How NO is generated in nitrogen-starved C. reinhardtii (i.e., in the absence of any external sources of nitrogen) remains to be elucidated. There are two well identified intracellular sources of NO (reviewed in Besson-Bard et al., 2008; Wilson et al., 2008; Gupta et al., 2011). The first one is l-Arg, converted into NO and l-citrulline by the nitric oxide synthase (NOS). This is the main NO-producing pathway in animals, but its relevance in plants is still a matter of debate (reviewed in Zemojtel et al., 2006; Fröhlich and Durner, 2011). To date, a NOS homolog has been found in the green alga Ostreococcus (Foresi et al., 2010) but not in other algae nor in land plants. Furthermore l-Arg plays little role in NO production under nitrogen replete conditions, whether in C. reinhardtii (Sakihama et al., 2002) or in the closely related alga Scenedesmus obliquus (Mallick et al., 2000b). Nevertheless, the NO-dependent repression of the nitrate assimilation pathway by ammonium is partially prevented upon addition of the l-Arg analog l-NAME, a known NOS inhibitor, suggesting some NOS-like activity in C. reinhardtii (de Montaigu et al., 2010). In any event, we could not test the effect of l-NAME on the loss of the cytochrome b6f complex during nitrogen starvation because the drug was efficiently used as a source of nitrogen by C. reinhardtii (data not shown).
Reduction of nitrite thus seems to be the major source of NO in plants (Mallick et al., 1999, 2000a). NO is produced by a variety of pathways identified in several recent studies (reviewed in Gupta et al., 2011), competed by the reduction of nitrite to ammonium by NiR. These pathways are either associated with the plasma membrane (Stöhr et al., 2001; Eick and Stöhr, 2012) or localized in peroxisomes (Barroso et al., 1999; Corpas et al., 2001, 2009; Del Río, 2011), mitochondria (Tischner et al., 2004; Gupta et al., 2010; Gupta and Igamberdiev, 2011), chloroplasts (Jasid et al., 2006; Galatro et al., 2013; Tewari et al., 2013), or the cytosol. The cytosolic NaR appears to be critical for the nitrite-dependent formation of NO since it provides the substrate to other nitrite-dependent NO-producing pathways by catalyzing the reduction of nitrate to nitrite, in addition to its intrinsic nitrite to NO reduction activity under hypoxic conditions (Dean and Harper, 1988; Yamasaki and Sakihama, 2000; reviewed in Meyer et al., 2005).
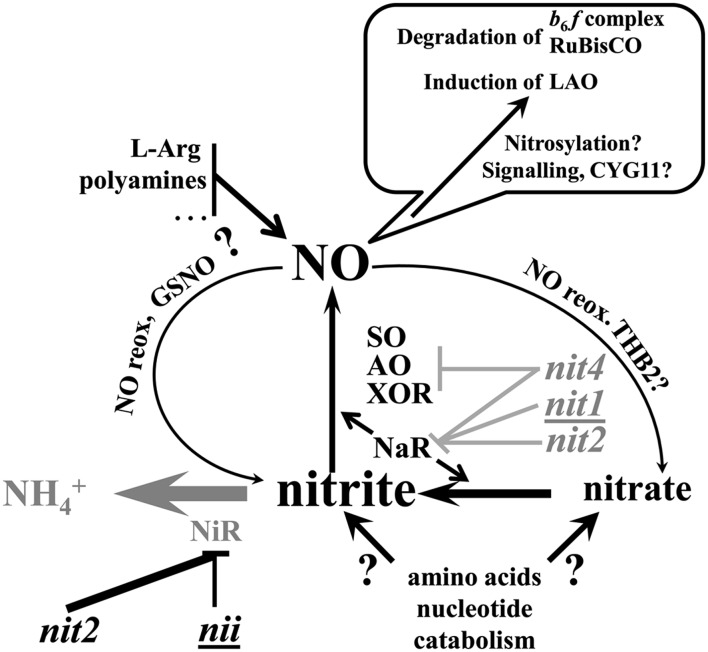
A number of our experiments support a predominant role of intracellular nitrite to NO production, as illustrated on the summary scheme shown Figure 13. Addition of nitrite to NiR mutants was as efficient at accelerating the degradation of the cytochrome b6f complex as was the addition of an NO donor to our reference strain WT-S24. Furthermore, the kinetics and amplitude of the degradation of the cytochrome b6f complex correlate well with the predicted size of the nitrite pool in different genetic backgrounds. Nitrite accumulation should be very low in nit1 or nit4 mutants showing hampered nitrite production in the absence of NaR, while nitrite is still consumed by NiR. Accordingly, nit1 cells accumulate very little NO, as seen by confocal microscopy. Nitrite pools should increase in the wild type where, in spite of an active NiR, NaR is also active. In nit2 strains lacking both NiR and NaR activities, nitrite produced by NaR-independent pathways may accumulate. It is of note that in the normoxic conditions required to observe the degradation of the cytochrome b6f during nitrogen starvation, NO can be converted back to nitrite as a result of the chemistry of the GSNO:GSH system (Singh et al., 1996), as well as spontaneously by reacting with ambient O2 or because of the NO oxidase activity of truncated hemoglobins. Among the 12 truncated hemoglobins found in C. reinhardtii (Hemschemeier et al., 2013a), THB2 is specifically induced more than 20-fold at the transcriptional level during nitrogen starvation (Sabeeha Merchant, personal communication). Thus, the status of intracellular nitrite and NO are intimately linked as part of a metabolic cycle and nitrite could be considered as a stable reservoir for NO production, at least in the absence of the NiR sink.
Figure 13.
Tentative Model for the Production of NO during Nitrogen Starvation.
As indicated in the balloon, NO contributes to the loss of the cytochrome b6f complex by tagging cytochrome b6f subunits and related proteins for degradation and/or by participating in a signal transduction pathway leading to gene activation and degradation of the cytochrome b6f complex. Enzymes or genetic contexts that favor the accumulation of nitrite and the production of NO are shown in black, while those counteracting these processes are shown in gray. Structural genes for the key enzymes of nitrogen metabolism are underlined. Because nit2 mutation has a dual effect, favoring nitrite accumulation in the absence of NiR, but preventing the reduction of nitrate to nitrite and of nitrite to NO in the absence of NaR, it appears in both colors. THB2 and CYG11 code for a truncated hemoglobin and for a NO-responsive guanylate cyclase, respectively, whose transcription is induced during nitrogen starvation.
Based on our results, NaR activity is required in the presence of NiR to keep the concentration of nitrite high enough for NO production. Yet, the phenotype of nit2 cells shows that the NaR catalytic activity per se is not required for productive NO synthesis during nitrogen starvation, as was also observed in anoxic conditions (Hemschemeier et al., 2013a, 2013b). NaR activity appears also dispensable for nitrite-dependent NO synthesis in Scenedesmus (Mallick et al., 1999) but seems required for NO evolution from nitrite by C. reinhardtii cells grown in nitrogen-replete conditions (Sakihama et al., 2002). However, NO production by NaR-defective mutants has been documented during hypoxia (Hemschemeier et al., 2013a) in mastoparan-treated cells (Yordanova et al., 2010) or in iron-limited cultures exposed to CO (Liping et al., 2013). Together, these results point to the prevalence of alternative NO-producing pathways in C. reinhardtii, probably coupled to the increased catabolism of amino acids and nucleotides undergone by nitrogen-starved cells. For instance, in many organisms, including vascular plants, xanthine oxidase-dehydrogenase (XOR), aldehyde oxidase (AO), and sulfite oxidase (SO), all MoCo-requiring enzymes, can reduce nitrite to NO (Tewari et al., 2009; B.L. Wang et al., 2010; Maia and Moura, 2011), even under normoxia, when using NADH as a substrate (Li et al., 2004). In that respect, we noted that the induction of LAO1, which displays the same regulation pattern as the degradation of cytochrome b6f and Rubisco, as it is much delayed and limited in genetic or environmental contexts that prevent the degradation of these proteins, was even more delayed in strain nit4-104 lacking the four enzymes NaR, AO, XOR, and SO, because of the absence of MoCo, than in strain nit1-137 lacking NaR only. Interestingly, both XOR and AO are upregulated about 5-fold at the mRNA level during nitrogen starvation (Miller et al., 2010; Sabeeha Merchant, personal communication), probably to cope with the increased purine catabolism resulting from the extensive mobilization of RNA and chloroplast DNA.
The Production of NO Is Part of a Signaling Pathway That Controls the Degradation of Cytochrome b6f and Biogenesis Factors
At this stage, we cannot yet assess properly the respective contributions to the loss of the cytochrome b6f complex of (1) protein posttranslational modifications (nitrosylation) that would target them for degradation and (2) activation of a NO signal transduction pathway. NO has been involved in a number of signaling pathways in C. reinhardtii, including acclimation to anaerobiosis (Hemschemeier et al., 2013a), iron homeostasis (Liping et al., 2013), cell death (Yordanova et al., 2010), copper stress (Zhang et al., 2008), and regulation of nitrogen assimilation (de Montaigu et al., 2010; Sanz-Luque et al., 2013). Here, the coregulation of cytochrome b6f degradation and nucleus-encoded LAO1 induction upon nitrogen starvation, both being slowed in nit1 mutants and accelerated in nit2 mutants, suggests that NO acts as a signaling molecule at least for the transcriptional induction of LAO1. Actually, NO bubbling or SNP addition proved insufficient by themselves to trigger the degradation of the cytochrome b6f complex in anoxic or phototrophic conditions in absence of nitrogen (data not shown). These observations suggest the recruitment of other components in a signaling pathway, besides protein nitrosylation, to activate the degradation of cytochrome b6f complex and biogenesis factors, as is typical in many stress-induced responses. This signaling pathway may include the nitrogen starvation–induced transcription factor NITROGEN RESPONSE REGULATOR1 (Boyle et al., 2012) or the chloroplast ortholog of the PII protein (Hsieh et al., 1998; Ermilova et al., 2013), which in (cyano)-bacteria signals the nitrogen status under antagonistic regulation by α-ketoglutarate (reviewed in Ninfa and Jiang, 2005). It may also involve CYG11, one of the NO-responsive guanylate cyclases that were found in the nuclear genome of C. reinhardtii (de Montaigu et al., 2010), whose expression is increased 20-fold during nitrogen starvation (Sabeeha Merchant, personal communication). Last, the signaling pathway may include some adaptor proteins of proteases that recognize and bind both the substrates for degradation and the chaperone subunit of the protease (reviewed in Kirstein et al., 2009). For instance, in cyanobacteria, the NblA adaptor mediates the proteolytic degradation of phycobilisomes during nitrogen or sulfur starvation because of its double affinity for phycobilisome rods and for ClpC (Collier and Grossman, 1994; Karradt et al., 2008).
This study shows how the extensive response of C. reinhardtii to N starvation is finely tuned by genetic and metabolic factors, some of which have been largely ignored thus far. As NO regulates photosynthesis shutdown that has deep implications for the intracellular allocation of carbon, it becomes critical to understand its signaling effects and to manipulate its production for better harnessing microalgae for biofuel production. A systematic study of nitrosylated proteins (focusing on cytochrome b6f subunits and biogenesis factors and Rubisco) in the experimental conditions or genetic contexts that do or do not lead to cytochrome b6f degradation upon nitrogen starvation will help identify the mechanisms that target proteins to proteolysis, while looking for changes in gene expression in the same conditions should provide clues to the signal transduction pathway.
METHODS
Strains, Media, Culture Conditions, and Chemicals
Wild-type and mutant strains (listed in Table 1) of Chlamydomonas reinhardtii were grown on a rotary shaker (120 rpm) in TAP medium, pH 7.2 (Harris, 1989) under continuous light (5 to 10 µE⋅m−2⋅s−1), except for the dum22 mutant that was grown in the same conditions but under 75 µE⋅m−2⋅s−1. For nitrogen starvation experiments, cells pregrown either in TAP or in minimum medium (4.1 mM K2HPO4, 2.65 mM KH2PO4, 0.3 mM CaCl2, 0.4 mM MgSO4, and 1/1000 Huntner trace solution) without bubbling of ambient air nor extra CO2 up to the mid log phase (2 × 106 cells mL−1) were centrifuged at 3000g for 5 min, washed once in nitrogen-free medium (Harris, 1989), resuspended at 2 × 106 cells mL−1 in nitrogen-free medium (150 mL in a 500-mL Erlenmeyer), and kept on a rotary shaker with vigorous aeration (225 rpm) to ensure a good aeration. Strains mH ftsh1-1.2+ and mH clpP-AUU.231+ were generated by crossing, according to Harris (1989), the strain mH mt− (Boulouis et al., 2011) with strains ftsH1-1 mt+ (Malnoë et al., 2014) and clpP-AUU mt+ (Majeran et al., 2000) and selecting the desired progeny by appropriate screens. Strain ptox2 {ΔpetB} was obtained by crossing the ptox2 mt− strain (Houille-Vernes et al., 2011) with strain {ΔpetB} mt+ (Kuras and Wollman, 1994). cPTIO, SNP, and DAF-FM DA were purchased from Sigma-Aldrich, cPTIO was purchased from Invitrogen Life Technologies, and GSNO was obtained from BioVision. GSNO solutions were prepared extemporaneously, and GSNO concentration was determined spectrophotometrically using molar extinction coefficient of 920 M−1⋅cm−1 at 335 nm.
Protein Preparation, Separation, and Analysis
Protein isolation, separation, and immunoblot analyses were performed as described (Kuras and Wollman, 1994), except that, for detection of the CCB1-4, CSS1, and CSS5 factors, proteins were transferred onto polyvinylidene difluoride rather than onto nitrocellulose membranes. Cell extracts were loaded on an equal chlorophyll basis. At least three biological replicas were performed for each experiment. Proteins were detected by ECL. Primary antibodies, diluted 100,000-fold (antibodies against cytochrome f, LAO1, D1, LHCII, and PsaA), 50,000-fold (CF1β and RbcL), 10,000-fold (cytochrome b6, Rieske, cytochrome b6f subunit IV, and NDA2), 5000-fold (CCS5, PTOX2, and PETO), 2500-fold (CCB3 and CCS1), or 300-fold (CCB1,2,4) were revealed by an horseradish peroxidase–conjugated antibody against rabbit IgG (Promega). Antibodies against D1, PsaA, and Rubisco LS were purchased from Agrisera. Other antibodies were described previously: cytochrome b6f subunits (Kuras and Wollman, 1994), ATP synthase subunit β (Drapier et al., 1992), CCBs (Kuras et al., 2007), CCS1 (Dreyfuss et al., 2003), CCS5 (Gabilly et al., 2010), PTOX2 (Houille-Vernes et al., 2011), and NDA2 (Desplats et al., 2009). TCA1-Flag and MCA1-HA were detected by ECL using monoclonal antibodies against the tags anti-Flag M2 (Sigma-Aldrich), anti HA.11 (Covance), and horseradish peroxidase–conjugated antibody against mouse IgG (Promega). Protein accumulation (normalized to that of the D1 subunit of PSII as internal standard) was, when required, quantified from ChemiDoc XRS+ (Bio-Rad) scans of the membrane, using the ImageLab (v3.0) software. Cytochrome f accumulation, normalized to that of the CF1β subunit as an internal standard, was quantified from phosphor imager (Molecular Dynamics) scans of immunoblots revealed with 125I protein A, using the ImageQuant software, as described (Choquet et al., 2003).
RNA Isolation and Analysis
RNA extraction and RNA gel blot analysis were performed as described (Drapier et al., 2002) with probes derived from coding sequences (Eberhard et al., 2002). When preparing the RNA templates for quantitative RT-PCR experiments, AIA was omitted from the lysis buffer. After extraction, 40 µg of RNA was resuspended in 200 μL of DNaseI buffer and treated at 37°C with DNase I (New England Biolabs; 25 units) before being further purified using the Qiagen RNeasy MinElute Cleanup Kit according to the manufacturer’s instruction. Reverse transcription was done on 1 µg of total RNA with the SuperScript III first-strand synthesis SuperMix for quantitative RT-PCR kit (Invitrogen) according to the manufacturer’s protocol using the random primers included in the kit. Quantitative PCR was performed in a GeneRotor 3000 apparatus (Corbet, Qiagen) in a final 25-μL reaction using the FASTStart SYBR Green Master Mix (Roche Applied Science), 0.5 μL (out of 21 µL) of the reverse transcriptase reaction mixture, and the primers listed in Supplemental Table 3, with the following cycling parameters: 95°C for 10’ (95°C for 10’’, 64°C for 15’’ to 72°C for 25’’) repeated 40 times. Fluorescence was recorded at the end of the elongation step. The specificity of PCR amplification was checked by a melting curve program (65 to 97°C with heating rate of 1°C per min and continuous fluorescence measurements). Absence of amplification from genomic DNA was checked by gel agarose electrophoresis. Results were analyzed using Rotor-Gene Q software (Qiagen), and data are expressed as relatives values with respect to the NAC2 mRNA level used as internal standard as this factor remains unaffected during nitrogen starvation (Raynaud et al., 2007).
Fluorescence Measurements
Fluorescence of liquid cultures, dark-adapted under strong agitation in open Erlenmeyer flasks for 30 min, was measured using a home-built fluorimeter.
Measurement of Reoxidation Kinetics of the PQ Pool
{ΔpetB} single mutant and ptox2 {ΔpetB} double mutant, starved for nitrogen for 34 h or grown in nitrogen-replete conditions, were collected by centrifugation and resuspended in 20 mM HEPES, pH 7.2, plus 20% Ficoll to avoid sedimentation. The in vivo rate of reoxidation of the plastoquinol pool in the dark was measured and calculated according to Houille-Vernes et al. (2011). In brief, the PSII photochemical rate and the variable part of fluorescence are linearly correlated. The area over the fluorescence rise curve represents the size of the pool of PSII electron acceptors. In mutants devoid of the cytochrome b6f complex, this area reflects the number of oxidized quinones before illumination (Bennoun, 1982). In the presence of DCMU, this area corresponds to one electron (transferred to QA) and can be used for data normalization. Following the full reduction of the PQ pool by continuous illumination, measurements performed after increasing periods of darkness allow analysis of the kinetics of plastoquinol oxidation by PTOX (Bennoun, 2001).
Confocal Microscopy
Cells, deprived of nitrogen for the indicated times, were incubated 1 h in the presence of 5 mM DAF-FM DA, then washed and concentrated 20-fold by centrifugation in nitrogen-free medium and immediately imaged at room temperature with an inverted Zeiss LSM 710 laser scanning microscope equipped with a Plan Apochromat ×63/1.4 oil immersion objective and a motorized stage. Excitation was performed with a 488-nm argon laser (3% power), and emitted light was collected simultaneously on two different channels, between 493 and 599 nm and 647 and 797 nm, to separate signals arising from the NO sensor and endogenous chlorophyll, respectively. Images were obtained by 5 × 5 tile scanning, and zooms were performed on representative individual cells. Images were collected and treated with the Zen 2011 software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: cytochrome f, CAA51422.1; cytochrome b6, CAA51423; subunit IV, CAA51424; petC, X76299.1; petO, AF222893; RbcL, J01399.1; AtpB, M13704.1; PsaA, X05845.1; PsbA, CAA25670; OEE2, M15187.1; LAO1, CRU78797; PTOX2, XP_001703466; NDA2, XP_001703643; CCS1, CRU70999; ccs5, GU362093; ccb1, EF190472; CCB2, EF190473; CCB3, EF190474; CCB4, EF190475; TCA1, EF503563.1; MCA1, AF330231.1; ClpP, L28803.1; FtsH1, XM_001690837; NaR, AF203033; and NiR, Y08937.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Induction of PTOX2 and NDA2 Is Independent of the Incident Light.
Supplemental Figure 2. The Redistribution of the Cytochrome b6f Complex during State Transitions Does Not Contribute to Its Loss during Nitrogen Deprivation.
Supplemental Figure 3. The Degradation of the Cytochrome b6f Biogenesis Factors Does Not Result from a Cascade of Proteolytic Events.
Supplemental Figure 4. The Degradation of Cytochrome b6f Complex Subunits and Biogenesis Factors Is Independent from the Photosynthetic Activity of the Cells.
Supplemental Figure 5. The Loss of the Cytochrome b6f Complex Is Accelerated upon Multiple Addition of SNP.
Supplemental Figure 6. Confocal Imaging of a Chlamydomonas Wild-Type Cell Grown in the Presence of Acetate but Not Preincubated with the DAF-FM Fluorochrome.
Supplemental Figure 7. Complementation of the nit1-305 Mutation Restores the Degradation of the Cytochrome b6f Complex.
Supplemental Table 1. Strains Used for the Crosses and Experiments Presented as Supplemental Data.
Supplemental Table 2. Genetic Analysis of the Determinism of the Cytochrome b6f Loss in Nitrate Assimilation Mutants.
Supplemental Table 3. Oligonucleotides Used in This Work.
Supplementary Material
Acknowledgments
We thank X. Johnson, R. Dent, E. Fernandez, and P. Cardol for their kind gifts of the petE-1, ccs1-CAL28.01.07, and NiR-defective and nda2RNAi strains. We thank S. Merchant and P. Hamel for the antibodies against CCS1 and CCS5 and all members of the UMR7141 for stimulating discussions. We thank D. Drapier for critical reading of the article and S. Bujaldon for her help with figures and logo. This work was supported by the CNRS and Université Pierre et Marie Curie, Paris 06, Unité Mixte de Recherche 7141, by the European Community (“SunBioPath” Contract FP7-KBBE-2009-3-02), by Agence Nationale de la Recherche Contract ANR-12-BSV8-0011-01, and by the “Initiative d'Excellence” program from the French State (Grant “DYNAMO,” ANR-11-LABX-0011-01). B.D., D.S.-M., and A.B. were “Attaché(e)s Temporaires d’Enseignement et de Recherche” at Université Pierre et Marie Curie, Paris VI. L.W. was supported by a SunBioPath individual fellowship from the European Community (FP7-KBBE-2009-3-02).
AUTHOR CONTRIBUTIONS
Y.C. and F.-A.W. designed the research. L.W., B.D., A.G., L.H.-V., A.B., D.S.-M., A.M., F.R., C.d.V., O.V., and Y.C. performed research. L.W., B.D., A.G., L.H.-V., F.R., O.V., Y.C., and F.-A.W. analyzed data. L.W., Y.C., and F.-A.W. wrote the article.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- DCMU
3-(3,4-dichorophenyl)-1,1-dimethylurea
- PQ
plastoquinone
- SNP
sodium nitroprusside
- cPTIO
2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- GSNO
S-nitrosoglutathione
- DAF-FM DA
4-amino-5-methylamino-2',7'-difluoro-fluorescein diacetate
- TAP
Tris-acetate-phosphate
- NaR
nitrate reductase
- NiR
nitrite reductase
- MoCo
molybdopterin cofactor
- NOS
nitric oxide synthase
- XOR
xanthine oxidase-dehydrogenase
- AO
aldehyde oxidase
- SO
sulfite oxidase
Footnotes
Online version contains Web-only data.
References
- Angelo M., Hausladen A., Singel D.J., Stamler J.S. (2008). Interactions of NO with hemoglobin: From microbes to man. Methods Enzymol. 436: 131–168 [DOI] [PubMed] [Google Scholar]
- Astier J., Kulik A., Koen E., Besson-Bard A., Bourque S., Jeandroz S., Lamotte O., Wendehenne D. (2012). Protein S-nitrosylation: What’s going on in plants? Free Radic. Biol. Med. 53: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Astier J., Rasul S., Koen E., Manzoor H., Besson-Bard A., Lamotte O., Jeandroz S., Durner J., Lindermayr C., Wendehenne D. (2011). S-nitrosylation: An emerging post-translational protein modification in plants. Plant Sci. 181: 527–533 [DOI] [PubMed] [Google Scholar]
- Atteia A., de Vitry C., Pierre Y., Popot J.-L. (1992). Identification of mitochondrial proteins in membrane preparations from Chlamydomonas reinhardtii. J. Biol. Chem. 267: 226–234 [PubMed] [Google Scholar]
- Barroso J.B., Corpas F.J., Carreras A., Sandalio L.M., Valderrama R., Palma J.M., Lupiáñez J.A., del Río L.A. (1999). Localization of nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 274: 36729–36733 [DOI] [PubMed] [Google Scholar]
- Bennoun P. (2001). Chlororespiration and the process of carotenoid biosynthesis. Biochim. Biophys. Acta 1506: 133–142 [DOI] [PubMed] [Google Scholar]
- Bennoun P. (1982). Evidence for a respiratory chain in the chloroplast. Proc. Natl. Acad. Sci. USA 79: 4352–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A., Pugin A., Wendehenne D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Boulouis A., Raynaud C., Bujaldon S., Aznar A., Wollman F.A., Choquet Y. (2011). The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle N.R., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulté L., Bennoun P. (1990). Translational accuracy and sexual differentiation in Chlamydomonas reinhardtii. Curr. Genet. 18: 155–160 [DOI] [PubMed] [Google Scholar]
- Bulté L., Wollman F.A. (1992). Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6/f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur. J. Biochem. 204: 327–336 [DOI] [PubMed] [Google Scholar]
- Camargo A., Llamas A., Schnell R.A., Higuera J.J., González-Ballester D., Lefebvre P.A., Fernández E., Galván A. (2007). Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 19: 3491–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamizo-Ampudia A., Galvan A., Fernandez E., Llamas A. (2013). Characterization of Chlamydomonas 102 and 104 mutants reveals intermolecular complementation in the molybdenum cofactor protein CNX1E. Protist 164: 116–128 [DOI] [PubMed] [Google Scholar]
- Choquet Y., Vallon O. (2000). Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634 [DOI] [PubMed] [Google Scholar]
- Choquet Y., Zito F., Wostrikoff K., Wollman F.A. (2003). Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 15: 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J.L., Grossman A.R. (1994). A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F.J., Barroso J.B., del Río L.A. (2001). Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 6: 145–150 [DOI] [PubMed] [Google Scholar]
- Corpas F.J., Hayashi M., Mano S., Nishimura M., Barroso J.B. (2009). Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiol. 151: 2083–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., and Grossman, A. (1998). Responses to deficiencies in macronutrients. In The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas, M. Goldschmidt-Clermont, J.-D. Rochaix, S. Merchant, eds (Dordrecht/Boston/London: Kluwer Academic Publishers), pp. 603–635. [Google Scholar]
- Dean J.V., Harper J.E. (1988). The conversion of nitrite to nitrogen oxide(s) by the constitutive NAD(P)H-nitrate reductase enzyme from soybean. Plant Physiol. 88: 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Río L.A. (2011). Peroxisomes as a cellular source of reactive nitrogen species signal molecules. Arch. Biochem. Biophys. 506: 1–11 [DOI] [PubMed] [Google Scholar]
- de Montaigu A., Sanz-Luque E., Galván A., Fernández E. (2010). A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 22: 1532–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C., Mus F., Cuiné S., Billon E., Cournac L., Peltier G. (2009). Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J. Biol. Chem. 284: 4148–4157 [DOI] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Stern D.B., Wollman F.A. (2002). A dominant nuclear mutation in Chlamydomonas identifies a factor controlling chloroplast mRNA stability by acting on the coding region of the atpA transcript. Plant J. 31: 687–697 [DOI] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Wollman F.A. (1992). Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell 4: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss B.W., Hamel P.P., Nakamoto S.S., Merchant S. (2003). Functional analysis of a divergent system II protein, Ccs1, involved in c-type cytochrome biogenesis. J. Biol. Chem. 278: 2604–2613 [DOI] [PubMed] [Google Scholar]
- Eberhard S., Drapier D., Wollman F.-A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Ederli L., Reale L., Madeo L., Ferranti F., Gehring C., Fornaciari M., Romano B., Pasqualini S. (2009). NO release by nitric oxide donors in vitro and in planta. Plant Physiol. Biochem. 47: 42–48 [DOI] [PubMed] [Google Scholar]
- Eick M., Stöhr C. (2012). Denitrification by plant roots? New aspects of plant plasma membrane-bound nitrate reductase. Protoplasma 249: 909–918 [DOI] [PubMed] [Google Scholar]
- Ermilova E., Lapina T., Zalutskaya Z., Minaeva E., Fokina O., Forchhammer K. (2013). PII signal transduction protein in Chlamydomonas reinhardtii: Localization and expression pattern. Protist 164: 49–59 [DOI] [PubMed] [Google Scholar]
- Fernandez E., Galvan A. (2008). Nitrate assimilation in Chlamydomonas. Eukaryot. Cell 7: 555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresi N., Correa-Aragunde N., Parisi G., Caló G., Salerno G., Lamattina L. (2010). Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22: 3816–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich A., Durner J. (2011). The hunt for plant nitric oxide synthase (NOS): Is one really needed? Plant Sci. 181: 401–404 [DOI] [PubMed] [Google Scholar]
- Gabilly S.T., Dreyfuss B.W., Karamoko M., Corvest V., Kropat J., Page M.D., Merchant S.S., Hamel P.P. (2010). CCS5, a thioredoxin-like protein involved in the assembly of plastid c-type cytochromes. J. Biol. Chem. 285: 29738–29749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro A., Puntarulo S., Guiamet J.J., Simontacchi M. (2013). Chloroplast functionality has a positive effect on nitric oxide level in soybean cotyledons. Plant Physiol. Biochem. 66: 26–33 [DOI] [PubMed] [Google Scholar]
- Galvan A., Fernández E. (2001). Eukaryotic nitrate and nitrite transporters. Cell. Mol. Life Sci. 58: 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi M.L., Zhang L., Lee J.W., Flynn T., Seibert M., Greenbaum E., Melis A. (2000). Microalgae: A green source of renewable H(2). Trends Biotechnol. 18: 506–511 [DOI] [PubMed] [Google Scholar]
- González-Ballester D., Camargo A., Fernández E. (2004). Ammonium transporter genes in Chlamydomonas: The nitrate-specific regulatory gene Nit2 is involved in Amt1;1 expression. Plant Mol. Biol. 56: 863–878 [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Croft M., Gladyshev V.N., Merchant S.S., Posewitz M.C., Prochnik S., Spalding M.H. (2007). Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10: 190–198 [DOI] [PubMed] [Google Scholar]
- Gupta K.J., Fernie A.R., Kaiser W.M., van Dongen J.T. (2011). On the origins of nitric oxide. Trends Plant Sci. 16: 160–168 [DOI] [PubMed] [Google Scholar]
- Gupta K.J., Igamberdiev A.U. (2011). The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 11: 537–543 [DOI] [PubMed] [Google Scholar]
- Gupta K.J., Igamberdiev A.U., Kaiser W.M. (2010). New insights into the mitochondrial nitric oxide production pathways. Plant Signal. Behav. 5: 999–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel P., Olive J., Pierre Y., Wollman F.A., de Vitry C. (2000). A new subunit of cytochrome b6f complex undergoes reversible phosphorylation upon state transition. J. Biol. Chem. 275: 17072–17079 [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (2009). The genus Chlamydomonas In The Chlamydomonas Source Book, 2nd ed, E.H. Harris, ed (San Diego, CA: Academic Press, Elsevier), pp. 1–24. [Google Scholar]
- Hemschemeier A., Casero D., Liu B., Benning C., Pellegrini M., Happe T., Merchant S.S. (2013b). Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell 25: 3186–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A., Düner M., Casero D., Merchant S.S., Winkler M., Happe T. (2013a). Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc. Natl. Acad. Sci. USA 110: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houille-Vernes L., Rappaport F., Wollman F.A., Alric J., Johnson X. (2011). Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc. Natl. Acad. Sci. USA 108: 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M.H., Lam H.M., van de Loo F.J., Coruzzi G. (1998). A PII-like protein in Arabidopsis: Putative role in nitrogen sensing. Proc. Natl. Acad. Sci. USA 95: 13965–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Huang X., von Rad U., Durner J. (2002). Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923 [DOI] [PubMed] [Google Scholar]
- Jaba I.M., Zhuang Z.W., Li N., Jiang Y., Martin K.A., Sinusas A.J., Papademetris X., Simons M., Sessa W.C., Young L.H., Tirziu D. (2013). NO triggers RGS4 degradation to coordinate angiogenesis and cardiomyocyte growth. J. Clin. Invest. 123: 1718–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans F., Mignolet E., Houyoux P.A., Cardol P., Ghysels B., Cuiné S., Cournac L., Peltier G., Remacle C., Franck F. (2008). A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl. Acad. Sci. USA 105: 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasid S., Simontacchi M., Bartoli C.G., Puntarulo S. (2006). Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol. 142: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K., Inagawa T., Wilkinson A.J., Tatsuta T., Ogura T. (1999). Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem. 274: 26225–26232 [DOI] [PubMed] [Google Scholar]
- Karradt A., Sobanski J., Mattow J., Lockau W., Baier K. (2008). NblA, a key protein of phycobilisome degradation, interacts with ClpC, a HSP100 chaperone partner of a cyanobacterial Clp protease. J. Biol. Chem. 283: 32394–32403 [DOI] [PubMed] [Google Scholar]
- Kim S., Wing S.S., Ponka P. (2004). S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol. Cell. Biol. 24: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L., Schnell R.A., Fernández E., Lefebvre P.A. (1989). Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109: 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J., Molière N., Dougan D.A., Turgay K. (2009). Adapting the machine: Adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7: 589–599 [DOI] [PubMed] [Google Scholar]
- Kuras R., Saint-Marcoux D., Wollman F.A., de Vitry C. (2007). A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 104: 9906–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., Wollman F.A. (1994). The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.M., Kim B.Y., Li L., Morgan E.T. (2008). Nitric oxide-dependent proteasomal degradation of cytochrome P450 2B proteins. J. Biol. Chem. 283: 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Samouilov A., Liu X., Zweier J.L. (2004). Characterization of the effects of oxygen on xanthine oxidase-mediated nitric oxide formation. J. Biol. Chem. 279: 16939–16946 [DOI] [PubMed] [Google Scholar]
- Lien T., Schreiner Ø. (1975). Purification of a derepressible arylsulfatase from Chlamydomonas reinhardti: Properties of the enzyme in intact cells and in purified state. Biochim. Biophys. Acta Enzymol. 384: 168–179 [DOI] [PubMed] [Google Scholar]
- Liping Z., Hongbo S., Xiaohua L., Zhaopu L. (2013). Gene regulation of iron-deficiency responses is associated with carbon monoxide and heme oxydase 1 in Chlamydomonas reinhardtii. PLoS ONE 8: e53835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiselay C., Gumpel N.J., Girard-Bascou J., Watson A.T., Purton S., Wollman F.A., Choquet Y. (2008). Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 28: 5529–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth J., Noirel J., Pandhal J., Wright P.C., Vaidyanathan S. (2012). HILIC- and SCX-based quantitative proteomics of Chlamydomonas reinhardtii during nitrogen starvation induced lipid and carbohydrate accumulation. J. Proteome Res. 11: 5959–5971 [DOI] [PubMed] [Google Scholar]
- Maia L.B., Moura J.J. (2011). Nitrite reduction by xanthine oxidase family enzymes: A new class of nitrite reductases. J. Biol. Inorg. Chem. 16: 443–460 [DOI] [PubMed] [Google Scholar]
- Majeran, W. (2002). La Proteolyse dans le Chloroplaste de Chlamydomonas reinhardtii: Rôle et Organisation Structurale de la Protéase ClpP. PhD dissertation (Paris: Institut National Agronomique Paris-Grignon). [Google Scholar]
- Majeran W., Wollman F.A., Vallon O. (2000). Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick N., Mohn F.H., Rai L.C., Soeder C.J. (2000b). Evidence for the non-involvement of nitric oxide synthase in nitric oxide production by the green alga Scenedesmus obliquus. J. Plant Physiol. 156: 423–426 [Google Scholar]
- Mallick N., Mohn F.H., Soeder C.J. (2000a). Evidence supporting nitrite-dependent NO release by the green microalga Scenedesmus obliquus. J. Plant Physiol. 157: 40–46 [Google Scholar]
- Mallick N., Rai L.C., Mohn F.H., Soeder C.J. (1999). Studies on nitric oxide (NO) formation by the green alga Scenedesmus obliquus and the diazotrophic cyanobacterium Anabaena doliolum. Chemosphere 39: 1601–1610 [DOI] [PubMed] [Google Scholar]
- Malnoë A., Wang F., Girard-Bascou J., Wollman F.-A., de Vitry C. (2014). Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. Plant Cell 26: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N.C., Chiang K.S., Goodenough U.W. (1976). Turnover of chloroplast and cytoplasmic ribosomes during gametogenesis in Chlamydomonas reinhardi. Dev. Biol. 51: 190–201 [DOI] [PubMed] [Google Scholar]
- Martin N.C., Goodenough U.W. (1975). Gametic differentiation in Chlamydomonas reinhardtii. I. Production of gametes and their fine structure. J. Cell Biol. 67: 587–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., Helmann J.D. (2012). Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 60: 91–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C., Lea U.S., Provan F., Kaiser W.M., Lillo C. (2005). Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth. Res. 83: 181–189 [DOI] [PubMed] [Google Scholar]
- Miller R., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisse S., Zaffagnini M., Gao X.H., Lemaire S.D., Marchand C.H. (December 13, 2013). Insight into protein S-nitrosylation in Chlamydomonas reinhardtii. Antioxid. Redox Signal. http://dx.doi.org/10.1089/ars.2013.5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A., Mandon J., Cristescu S.M., Harren F.J., Prats E. (2011). Methods of nitric oxide detection in plants: A commentary. Plant Sci. 181: 509–519 [DOI] [PubMed] [Google Scholar]
- Navarro M.T., Guerra E., Fernández E., Galván A. (2000). Nitrite reductase mutants as an approach to understanding nitrate assimilation in Chlamydomonas reinhardtii. Plant Physiol. 122: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A.J., Jiang P. (2005). PII signal transduction proteins: Sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8: 168–173 [DOI] [PubMed] [Google Scholar]
- Peltier G., Schmidt G.W. (1991). Chlororespiration: An adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 88: 4791–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G., Happe T., Hemschemeier A. (2012). Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii. Planta 235: 729–745 [DOI] [PubMed] [Google Scholar]
- Plumley F.G., Schmidt G.W. (1989). Nitrogen-dependent regulation of photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 86: 2678–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada A., Galván A., Schnell R.A., Lefebvre P.A., Fernández E. (1993). Five nitrate assimilation-related loci are clustered in Chlamydomonas reinhardtii. Mol. Gen. Genet. 240: 387–394 [DOI] [PubMed] [Google Scholar]
- Quesada A., Gómez I., Fernández E. (1998). Clustering of the nitrite reductase gene and a light-regulated gene with nitrate assimilation loci in Chlamydomonas reinhardtii. Planta 206: 259–265 [DOI] [PubMed] [Google Scholar]
- Quisel J.D., Wykoff D.D., Grossman A.R. (1996). Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 111: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.A., Choquet Y. (2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 104: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C., Cardol P., Coosemans N., Gaisne M., Bonnefoy N. (2006). High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 103: 4771–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakihama Y., Nakamura S., Yamasaki H. (2002). Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: An alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol. 43: 290–297 [DOI] [PubMed] [Google Scholar]
- Sanz-Luque E., Ocaña-Calahorro F., Llamas A., Galvan A., Fernandez E. (2013). Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii. J. Exp. Bot. 64: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Schreiner Ø., Lien T., Knutsen G. (1975). The capacity for arylsulfatase synthesis in synchronous and synchronized cultures of Chlamydomonas reinhardti. Biochim. Biophys. Acta Enzymol. 384: 180–193 [DOI] [PubMed] [Google Scholar]
- Sears B.B., Boynton J.E., Gillham N.W. (1980). The effect of gametogenesis regimes on the chloroplast genetic system of Chlamydomonas reinhardti. Genetics 96: 95–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylidès C., Li-Beisson Y., Peltier G. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersma P.W., Chiang K.S. (1971). Conservation and degradation of cytoplasmic and chloroplast ribosomes in Chlamydomonas reinhardtii. J. Mol. Biol. 58: 167–185 [DOI] [PubMed] [Google Scholar]
- Singh S.P., Wishnok J.S., Keshive M., Deen W.M., Tannenbaum S.R. (1996). The chemistry of the S-nitrosoglutathione/glutathione system. Proc. Natl. Acad. Sci. USA 93: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza J.M., Choi I., Chen Q., Weisse M., Daikhin E., Yudkoff M., Obin M., Ara J., Horwitz J., Ischiropoulos H. (2000). Proteolytic degradation of tyrosine nitrated proteins. Arch. Biochem. Biophys. 380: 360–366 [DOI] [PubMed] [Google Scholar]
- Stöhr C., Strube F., Marx G., Ullrich W.R., Rockel P. (2001). A plasma membrane-bound enzyme of tobacco roots catalyses the formation of nitric oxide from nitrite. Planta 212: 835–841 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Sato N., Tsuzuki M. (2007). Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett. 581: 4519–4522 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Tsuzuki M., Sato N. (2010). Regulation of synthesis and degradation of a sulfolipid under sulfur-starved conditions and its physiological significance in Chlamydomonas reinhardtii. New Phytol. 185: 676–686 [DOI] [PubMed] [Google Scholar]
- Tang C.H., Wei W., Liu L. (2012). Regulation of DNA repair by S-nitrosylation. Biochim. Biophys. Acta 1820: 730–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.D., Miranda K.M., Colton C.A., Citrin D., Espey M.G., Wink D.A. (2003). Heme proteins and nitric oxide (NO): The neglected, eloquent chemistry in NO redox signaling and regulation. Antioxid. Redox Signal. 5: 307–317 [DOI] [PubMed] [Google Scholar]
- Terauchi A.M., Peers G., Kobayashi M.C., Niyogi K.K., Merchant S.S. (2010). Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 105: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R.K., Kumar P., Kim S., Hahn E.J., Paek K.Y. (2009). Nitric oxide retards xanthine oxidase-mediated superoxide anion generation in Phalaenopsis flower: An implication of NO in the senescence and oxidative stress regulation. Plant Cell Rep. 28: 267–279 [DOI] [PubMed] [Google Scholar]
- Tewari R.K., Prommer J., Watanabe M. (2013). Endogenous nitric oxide generation in protoplast chloroplasts. Plant Cell Rep. 32: 31–44 [DOI] [PubMed] [Google Scholar]
- Tischner R., Planchet E., Kaiser W.M. (2004). Mitochondrial electron transport as a source for nitric oxide in the unicellular green alga Chlorella sorokiniana. FEBS Lett. 576: 151–155 [DOI] [PubMed] [Google Scholar]
- Toepel J., Albaum S.P., Arvidsson S., Goesmann A., la Russa M., Rogge K., Kruse O. (2011). Construction and evaluation of a whole genome microarray of Chlamydomonas reinhardtii. BMC Genomics 12: 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O., Bulté L., Kuras R., Olive J., Wollman F.A. (1993). Extensive accumulation of an extracellular L-amino-acid oxidase during gametogenesis of Chlamydomonas reinhardtii. Eur. J. Biochem. 215: 351–360 [DOI] [PubMed] [Google Scholar]
- Wang B.L., Tang X.Y., Cheng L.Y., Zhang A.Z., Zhang W.H., Zhang F.S., Liu J.Q., Cao Y., Allan D.L., Vance C.P., Shen J.B. (2010). Nitric oxide is involved in phosphorus deficiency-induced cluster-root development and citrate exudation in white lupin. New Phytol. 187: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen C., Loake G.J., Chu C. (2010). Nitric oxide: Promoter or suppressor of programmed cell death? Protein Cell 1: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. (2009). Algal lipid bodies: Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8: 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Yang Z., Tang C.H., Liu L. (2011). Targeted deletion of GSNOR in hepatocytes of mice causes nitrosative inactivation of O6-alkylguanine-DNA alkyltransferase and increased sensitivity to genotoxic diethylnitrosamine. Carcinogenesis 32: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffels R.H., Barbosa M.J. (2010). An outlook on microalgal biofuels. Science 329: 796–799 [DOI] [PubMed] [Google Scholar]
- Wilson I.D., Neill S.J., Hancock J.T. (2008). Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 31: 622–631 [DOI] [PubMed] [Google Scholar]
- Wollman F.A., Minai L., Nechushtai R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim. Biophys. Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Work V.H., Radakovits R., Jinkerson R.E., Meuser J.E., Elliott L.G., Vinyard D.J., Laurens L.M., Dismukes G.C., Posewitz M.C. (2010). Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot. Cell 9: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Choquet Y., Wollman F.A., Girard-Bascou J. (2001). TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff D.D., Davies J.P., Melis A., Grossman A.R. (1998). The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.J., Shen W.B. (2012). In vivo imaging of nitric oxide and reactive oxygen species using laser scanning confocal microscopy. Methods Mol. Biol. 913: 191–200 [DOI] [PubMed] [Google Scholar]
- Xie Z., Culler D., Dreyfuss B.W., Kuras R., Wollman F.A., Girard-Bascou J., Merchant S. (1998). Genetic analysis of chloroplast c-type cytochrome assembly in Chlamydomonas reinhardtii: One chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka G., Glazer A.N. (1980). Dynamic aspects of phycobilisome structure-Phycobilisome turnover during nitrogen starvation in Synechococcus sp. Arch. Microbiol. 124: 39–47 [Google Scholar]
- Yamasaki H., Sakihama Y. (2000). Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 468: 89–92 [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff S., Zimmer S.L., Komine Y., Stern D.B. (2007). Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell 19: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanova Z.P., Iakimova E.T., Cristescu S.M., Harren F.J., Kapchina-Toteva V.M., Woltering E.J. (2010). Involvement of ethylene and nitric oxide in cell death in mastoparan-treated unicellular alga Chlamydomonas reinhardtii. Cell Biol. Int. 34: 301–308 [DOI] [PubMed] [Google Scholar]
- Zemojtel T., Fröhlich A., Palmieri M.C., Kolanczyk M., Mikula I., Wyrwicz L.S., Wanker E.E., Mundlos S., Vingron M., Martasek P., Durner J. (2006). Plant nitric oxide synthase: A never-ending story? Trends Plant Sci. 11: 524–525, author reply 526–528 [DOI] [PubMed] [Google Scholar]
- Zhang L., Happe T., Melis A. (2002). Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214: 552–561 [DOI] [PubMed] [Google Scholar]
- Zhang L.P., Mehta S.K., Liu Z.P., Yang Z.M. (2008). Copper-induced proline synthesis is associated with nitric oxide generation in Chlamydomonas reinhardtii. Plant Cell Physiol. 49: 411–419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.