Figure 4.
Positional Cloning and Expression Analysis of GPA3.
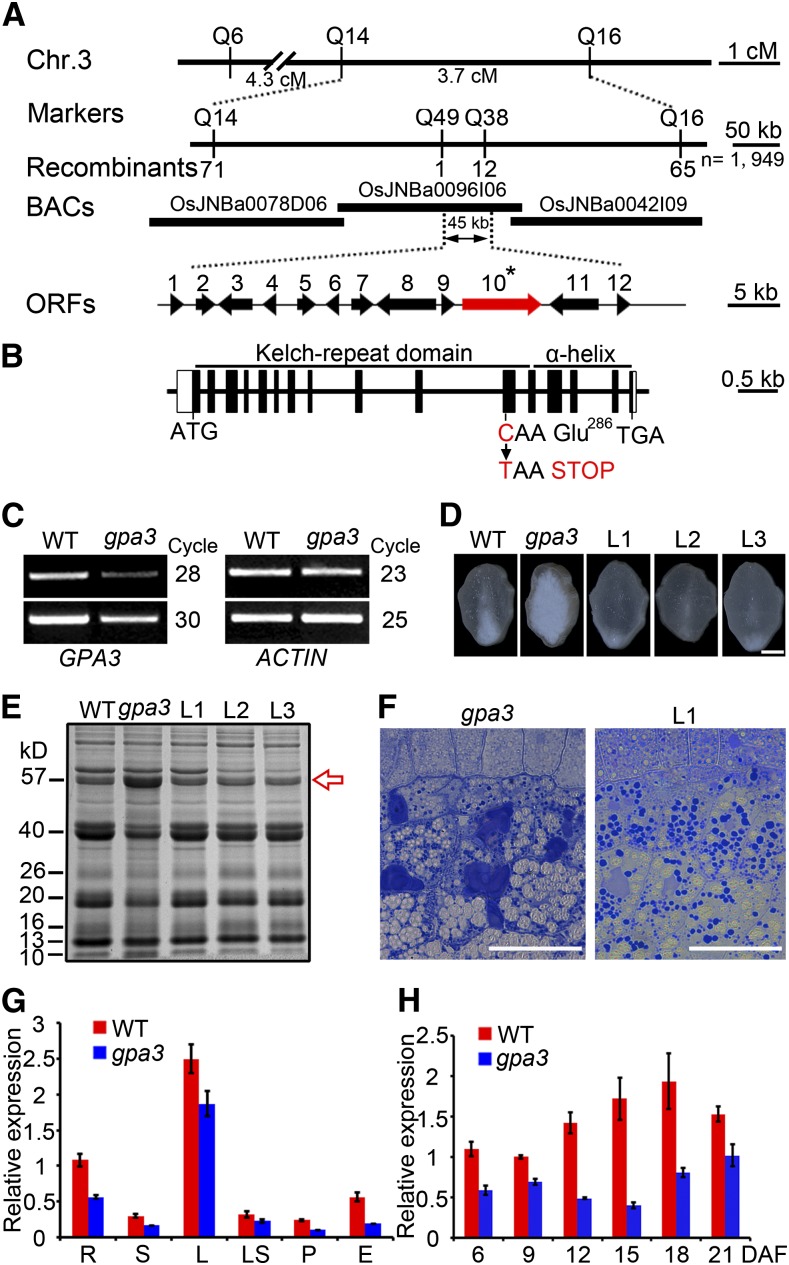
(A) Fine mapping of the GPA3 locus. The molecular markers and the number of recombinants are shown. cM, centimorgan; ORF, open reading frame.
(B) Exon/intron structure and mutation site of GPA3. The GPA3 gene comprises 16 exons (closed boxes) and 15 introns (open boxes). ATG and TGA represent the start and stop codons, respectively.
(C) RT-PCR assay showing that expression of the candidate GPA3 gene is reduced in the gpa3 mutant compared with the wild type.
(D) to (F) A 10.4-kb wild-type genomic segment of GPA3 completely rescues the grain appearance (D), the storage protein composition pattern (E), and the storage protein sorting defects (F) of the gpa3 mutant. L1 to L3 denote grains from three different T1 transgenic lines. The red arrow indicates the 57-kD proglutelins. Bar in (D) = 1 mm; bars in (F) = 50 μm.
(G) Real-time RT-PCR assay showing that GPA3 is expressed in various organs examined, with highest accumulation in the leaf. R, root; S, stem; L, leaf; LS, leaf sheath; P, panicle; E, endosperm.
(H) Real-time RT-PCR assay showing that GPA3 is expressed throughout endosperm development.
α-Tubulin was used as an internal control in real-time RT-PCR. For each RNA sample, three technical replicates were performed. Representative results from two biological replicates are shown. Values are means ± sd.